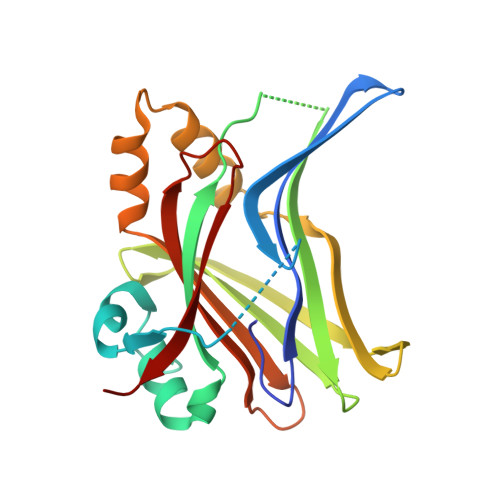
Discovery of reversible and covalent TEAD 1 selective inhibitors MSC-1254 and MSC-5046 based on one scaffold.
Carswell, E., Heinrich, T., Petersson, C., Gunera, J., Garg, S., Schwarz, D., Schlesiger, S., Fischer, F., Eichhorn, T., Calder, M., Smith, G., MacDonald, E., Wilson, H., Hazel, K., Trivier, E., Broome, R., Balsiger, A., Sirohi, S., Musil, D., Freire, F., Schilke, H., Dillon, C., Wienke, D.(2024) Bioorg Med Chem Lett 114: 129981-129981
- PubMed: 39369801
- DOI: https://doi.org/10.1016/j.bmcl.2024.129981
- Primary Citation of Related Structures:
9FZA - PubMed Abstract:
The Transcriptional Enhanced Associated Domain (TEAD) family of transcription factors are key components of the Hippo signalling family which play a crucial role in the regulation of cell proliferation, differentiation and apoptosis. The identification of inhibitors of the TEAD transcription factors are an attractive strategy for the development of novel anticancer therapies. A HTS campaign identified hit 1, which was optimised using structure-based drug design, to deliver potent TEAD1 selective inhibitors with both a reversible and covalent mode of inhibition. The preference for TEAD1 could be rationalised by steric differences observed in the lower pocket of the palmitoylation-site between subtypes, with TEAD1 having the largest available volume to accommodate substitution in this region.
- Cancer Research Horizons, Jonas Webb Building, Babraham Research Campus, Cambridge CB22 3AT, UK. Electronic address: ecarswell@exscientia.co.uk.
Organizational Affiliation: