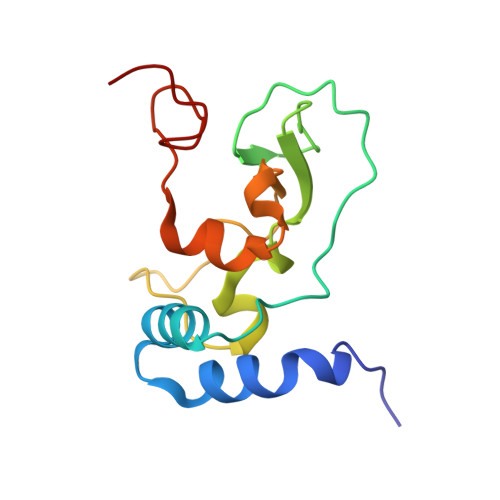
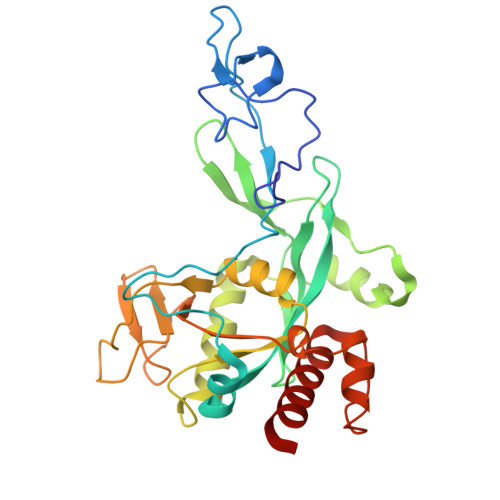
Structural basis for small molecule binding to the SARS-CoV-2 nsp10-nsp14 ExoN complex.
Kozielski, F., Fisher, S.Z., Ma, S., Al Busaidi, F., Krupinska, E., Nyblom, M., Sele, C., Sullivan, H.M., Krojer, T., Knecht, W.(2025) Nucleic Acids Res 53
- PubMed: 40794865
- DOI: https://doi.org/10.1093/nar/gkaf753
- Primary Citation of Related Structures:
9FW2, 9FWH, 9FWI, 9FWJ, 9FWK, 9FWL, 9FWM, 9FWN, 9FWO, 9FWP, 9FWQ, 9FWR, 9FWS, 9FWT, 9FWU, 9FZ4, 9FZK - PubMed Abstract:
Coronavirus outbreaks have occurred over the past 25 years with SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2) causing a global pandemic. The SARS-CoV-2 non-structural proteins 10 (nsp10) and 14 (nsp14) are considered as potential drug targets. Nsp10 stimulates the 3'-to-5' exoribonuclease (ExoN) activity of nsp14. The ExoN domain excises mis-incorporated nucleotides from the nascent RNA chain and therefore causes resistance to nucleoside analogue drugs. We crystallized the nsp10-nsp14 ExoN complex in distinct space groups, allowing us to describe conformational changes. In particular, the general base, His268, classifying the ExoN domain as a member of the DEDDh family, is trapped in the inactive and active orientations. By X-ray fragment screening, we identified five novel fragment binding sites in the nsp10-nsp14 interface, the hinge region connecting ExoN and N7-methyltransferase domains, and on nsp10. One new site in the nsp10-nsp14 interface accommodates nine structurally and chemically related hits, providing an initial structure-activity relationship study. We could also identify enantiomers of one fragment selectively bound to two different binding sites. The binding affinities of fragment hits were estimated using microscale thermophoresis and the new sites were investigated for their potential to inhibit protein-protein interactions between nsp10 and nsp14. Our fragments represent novel starting points for hit development by structure-based design.
- School of Pharmacy, University College London, 29-39 Brunswick Square, London WC1N 1AX, United Kingdom.
Organizational Affiliation: