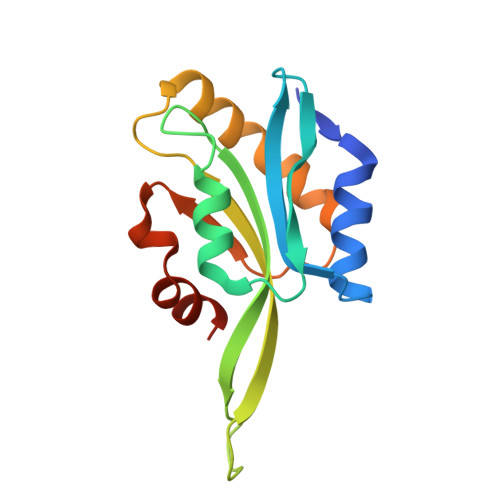
Crystal structure of Anopheles gambiae actin depolymerizing factor explains high affinity to monomeric actin.
Lasiwa, D., Kursula, I.(2025) FEBS J 292: 2381-2397
- PubMed: 39932036
- DOI: https://doi.org/10.1111/febs.70007
- Primary Citation of Related Structures:
9FP8 - PubMed Abstract:
Actin is an intrinsically dynamic protein, the function and state of which are modulated by actin-binding proteins. Actin-depolymerizing factors (ADF)/cofilins are ubiquitous actin-binding proteins that accelerate actin turnover. Malaria is an infectious disease caused by parasites of the genus Plasmodium, which belong to the phylum Apicomplexa. The parasites require two hosts to complete their life cycle: the definitive host, or the vector, an Anopheles spp. mosquito, and a vertebrate intermediate host, such as humans. Here, the malaria vector Anopheles gambiae ADF (AgADF) crystal structure is reported. AgADF has a conserved ADF/cofilin fold with six central β-strands surrounded by five α-helices with a long β-hairpin loop protruding out of the structure. The G- and F-actin-binding sites of AgADF are conserved, and the structure shows features of potential importance for regulation by membrane binding and redox state. AgADF binds monomeric ATP- and ADP-actin with a high affinity, having a nanomolar K d , and binds effectively also to actin filaments.
- Faculty of Biochemistry and Molecular Medicine, University of Oulu, Finland.
Organizational Affiliation: