Evaluating the impact of CRBN mutations on response to immunomodulatory drugs and novel cereblon E3 ligase modulators in myeloma.
Chrisochoidou, Y., Scarpino, A., Morales, S., Martin, S., Bird, S., Li, Y., Walker, B., Caldwell, J., Le Bihan, Y.V., Pawlyn, C.(2025) Blood 145: 2630-2644
- PubMed: 39841463
- DOI: https://doi.org/10.1182/blood.2024025861
- Primary Citation of Related Structures:
9FJX - PubMed Abstract:
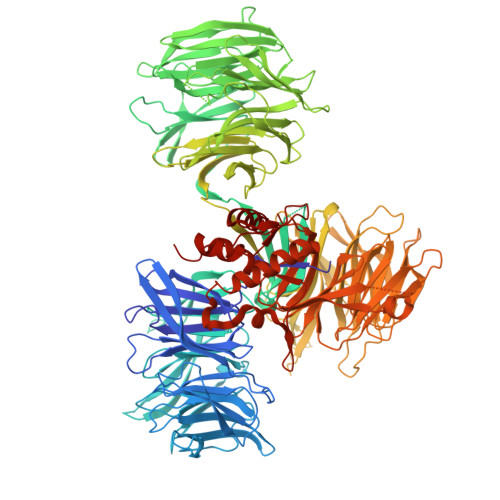
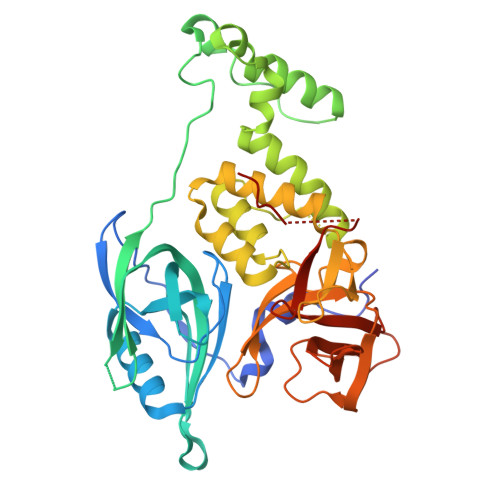
Immunomodulatory drug (IMiD) resistance is a key clinical challenge in myeloma treatment. Previous data suggests almost one third of myeloma patients acquire mutations in the key IMiD effector cereblon by the time they are pomalidomide refractory. Some events, including stop codons/frameshift mutations and copy loss, having clearly explicable effects on cereblon function. Missense mutations have also been reported throughout the length of cereblon but their functional impact has not been systematically studied. This study modelled selected missense mutations and examined their effect on cereblon function also analysing whether any mutations deleterious to IMiD action could be overcome using the novel cereblon binding agents (CELMoDs). Three patterns of response to missense mutations were apparent, mutations that led to complete loss of CRBN function for all agents, those that had no effect on CRBN function and those with agent-dependent effect on CRBN function. The latter group of 4 mutations were profiled in more detail with confirmatory experiments demonstrating an ability of the more potent CELMoDs to lead to neosubstrate degradation and cell death even though IMiDs were not active. Dynamic modelling based on a newly generated crystal structure of the DDB1/CRBN/lenalidomide complex, with greater resolution than those published to date, helped to understand the impact of these mutations. These results have important implications for the interpretation of CRBN sequencing results from patients for future therapy decisions, particularly differentiating those who may, despite relapsing on IMiDs with CRBN mutations, have the potential to still benefit from the use of CELMoD agents.
- The Institute of Cancer Research, London, United Kingdom.
Organizational Affiliation: