Identification of unique binding mode anti-NTF3 antibodies from a novel long VH CDR3 phage display library.
Chin, S.E., Gallego, P., Aagaard, A., Carmen, S., Barrett, N., Wolny, M., Cloarec, S., Paterson, J., Sivapalan, R., Hunt, J., Murray, T.V., Delaney, T., Sjogren, T., Neal, F.(2025) SLAS Discov 31: 100216-100216
- PubMed: 39832740
- DOI: https://doi.org/10.1016/j.slasd.2025.100216
- Primary Citation of Related Structures:
9FIK - PubMed Abstract:
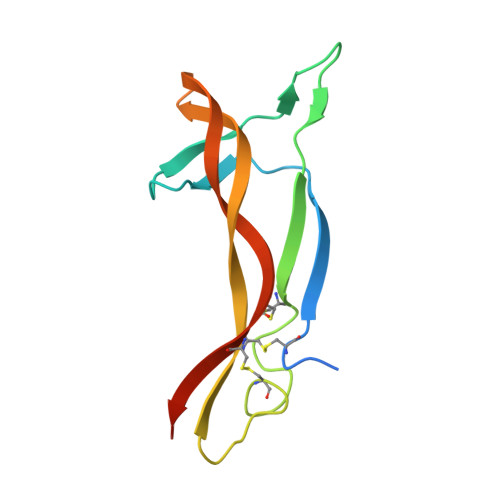
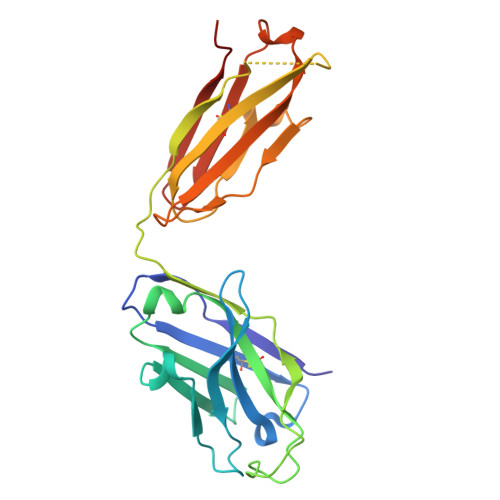
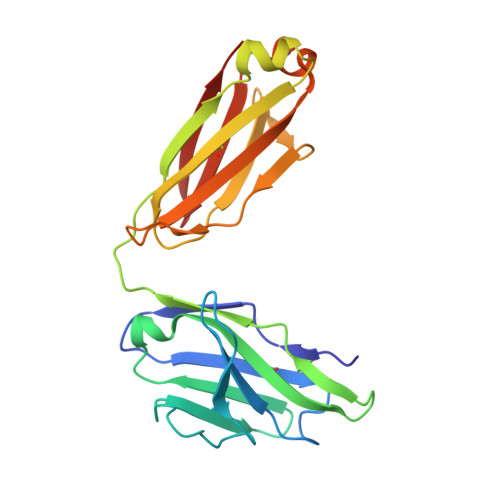
Neurotrophic factor 3 (NTF3) is a cysteine knot protein and a member of the nerve growth factor (NGF) family of cytokines. NTF3 engages the Trk family of receptor tyrosine kinases, playing a pivotal role in the development and function of both the central and peripheral nervous systems. Its involvement in neuronal survival, differentiation, and growth links NTF3 to a spectrum of neurodegenerative diseases. Consequently, targeting NTF3 with antibodies holds promise as a first in class therapeutic opportunity for a wide range of conditions. Specific and neutralizing antibodies against NTF3 were successfully isolated using phage display. Initial phage display selections revealed a preference of hits for a longer than average complementarity-determining region 3 (CDR3) in the heavy chain variable domain (VH). To investigate this further we developed a long loop length VH CDR3 antibody library that demonstrated increased hit rates versus a standard antibody library and allowed the isolation of IgG that demonstrated inhibition of functional activity, coupled with a favourable kinetic profile. Structural analysis of the Fab/NTF3 interaction, via X-ray crystallography, unveiled an unconventional interaction wherein regions beyond the longer CDR loops of the Fab induced ordering in a flexible loop on NTF3, which remained disordered in its free antigenic state. This comprehensive approach not only sheds light on the therapeutic potential of NTF3-specific antibodies but also provides critical structural details that enhance our understanding of the complex NTF3-Fab interaction thus offering valuable insights for future antibody design and therapeutic development.
- Biologics Engineering, Oncology R&D, AstraZeneca, Cambridge, UK.
Organizational Affiliation: