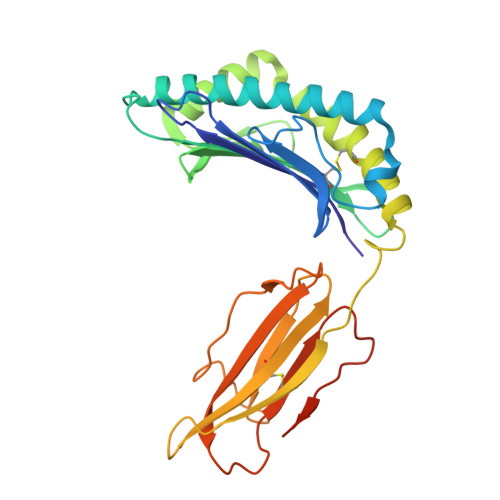
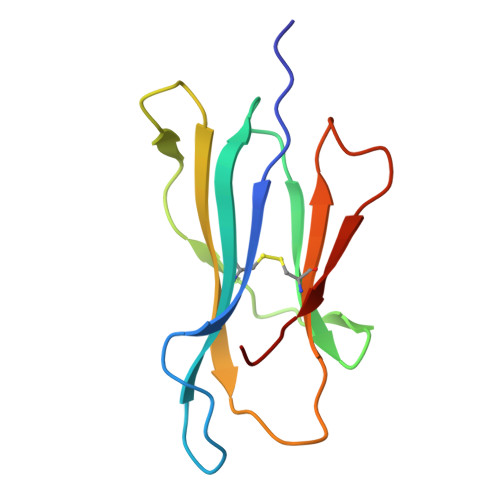
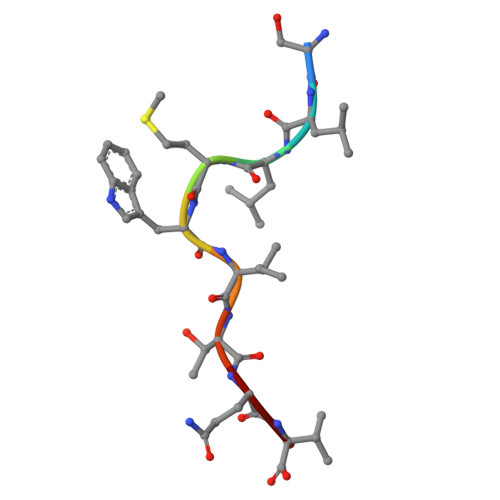
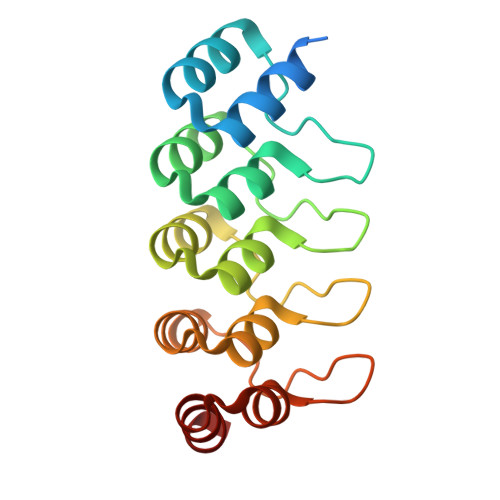
Development of DARPin T cell engagers for specific targeting of tumor-associated HLA/peptide complexes.
Venetz-Arenas, N., Schulte, T., Muller, S., Wallden, K., Fischer, S., Resink, T., Kadri, N., Paladino, M., Pina, N., Radom, F., Villemagne, D., Bruckmaier, S., Cornelius, A., Hospodarsch, T., Alici, E., Ljunggren, H.G., Chambers, B.J., Han, X., Sun, R., Carroni, M., Levitsky, V., Sandalova, T., Walser, M., Achour, A.(2025) iScience 28: 113926-113926
- PubMed: 41321628
- DOI: https://doi.org/10.1016/j.isci.2025.113926
- Primary Citation of Related Structures:
9EPA, 9FE1 - PubMed Abstract:
The balance between affinity and specificity in T cell receptor (TCR)-dependent targeting of HLA-restricted tumor-associated antigens presents a significant challenge for immunotherapy development. T cell engagers that circumvent these limitations are therefore of particular interest. We established a process to generate bispecific designed ankyrin repeat proteins (DARPins) that simultaneously target HLA-I/peptide complexes and CD3e. These high-affinity T cell engagers elicited CD8 + T cell activation against tumor targets with strong peptide specificity, as confirmed by X-scanning mutagenesis and functional killing assays. A cryo-EM structure of the ternary DARPin/HLA-A∗0201/NY-ESO1 157-165 complex revealed a rigid, concave DARPin surface spanning the full length of the peptide-binding cleft, contacting both α-helices and the peptide. The present findings reveal promising immuno-oncotherapeutic approaches and demonstrate the feasibility of rapidly developing DARPins with high affinity and specificity for HLA/peptide targets, which can be readily combined with a new generation of anti-CD3e-specific DARPins.
- Molecular Partners AG, Schlieren-Zurich, Switzerland.
Organizational Affiliation: