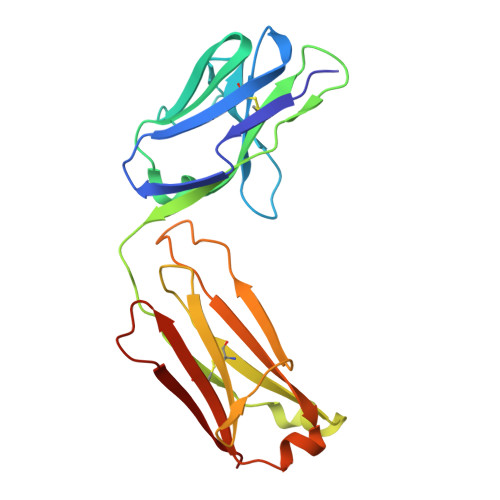
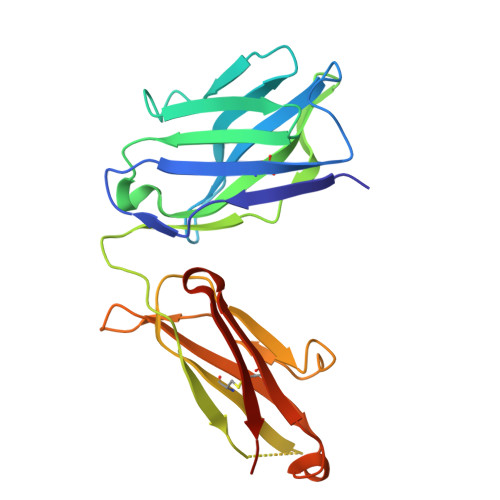
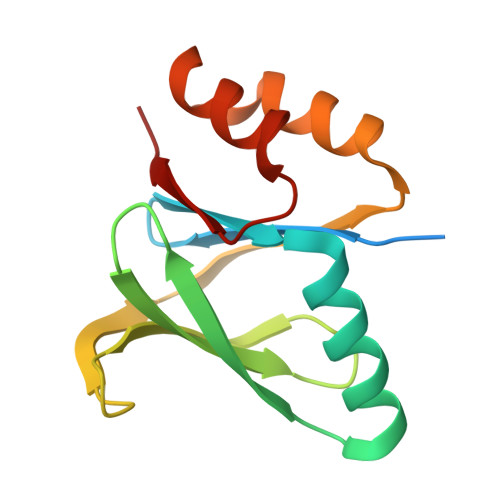
Accurate single-domain scaffolding of three nonoverlapping protein epitopes using deep learning.
Castro, K.M., Watson, J.L., Wang, J., Southern, J., Ayardulabi, R., Georgeon, S., Rosset, S., Baker, D., Correia, B.E.(2025) Nat Chem Biol
- PubMed: 41350440
- DOI: https://doi.org/10.1038/s41589-025-02083-z
- Primary Citation of Related Structures:
9F8Y, 9F90, 9F91 - PubMed Abstract:
De novo protein design has seen major success in scaffolding single functional motifs; however, in nature, most proteins present multiple functional sites. Here, we describe an approach to simultaneously scaffold multiple functional sites in a single-domain protein using deep learning. We designed small single-domain immunogens, under 130 residues, that present three distinct and irregular motifs from respiratory syncytial virus. These motifs together comprise nearly half of the designed proteins; hence, the overall folds are quite unusual with little global similarity to proteins in the Protein Data Bank. Despite this, X-ray crystal structures confirmed the accuracy of presentation of each of the motifs and the multiepitope design yields improved cross-reactive titers and neutralizing response compared to a single-epitope immunogen. The successful presentation of three distinct binding surfaces in a small single-domain protein highlights the power of generative deep learning methods to solve complex protein design problems.
- Institute of Bioengineering, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland.
Organizational Affiliation: