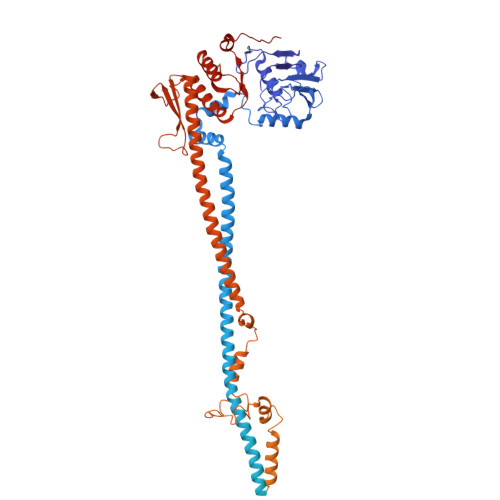
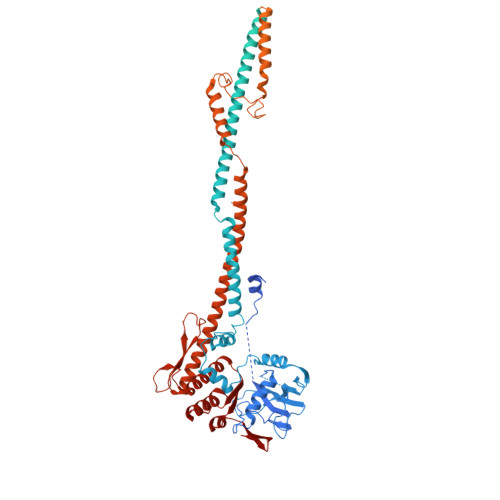
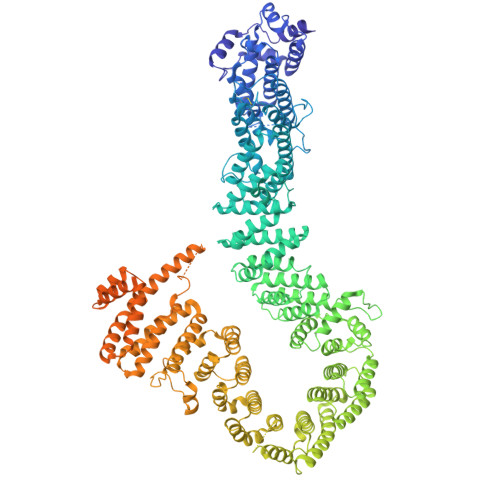
Condensin II activation by M18BP1.
Borsellini, A., Conti, D., Cutts, E.E., Harris, R.J., Walstein, K., Graziadei, A., Cecatiello, V., Aarts, T.F., Xie, R., Mazouzi, A., Sen, S., Hoencamp, C., Pleuger, R., Ghetti, S., Oberste-Lehn, L., Pan, D., Bange, T., Haarhuis, J.H.I., Perrakis, A., Brummelkamp, T.R., Rowland, B.D., Musacchio, A., Vannini, A.(2025) Mol Cell 85: 2688
- PubMed: 40614722
- DOI: https://doi.org/10.1016/j.molcel.2025.06.014
- Primary Citation of Related Structures:
9F5W - PubMed Abstract:
Condensin I and II promote the drastic spatial rearrangement of the human genome upon mitotic entry. While condensin II is known to initiate this process in early mitosis, what triggers its activation and loading onto chromatin at this juncture remains unclear. Through genetic and proteomic approaches, we identify MIS18-binding protein 1 (M18BP1), a protein required to maintain centromere identity, as the elusive factor required for condensin II localization to chromatin. M18BP1 directly binds condensin II's CAP-G2 subunit. The condensin II antagonist MCPH1 also binds to CAP-G2 and outcompetes M18BP1 during interphase to maintain the genome in its uncondensed state. A switch from MCPH1 to M18BP1 at mitotic onset activates condensin II, thus promoting proper chromosome condensation. Regulation of this M18BP1-condensin interaction thus determines both the uncondensed state of the interphase genome and its compacted state in mitosis.
- Human Technopole, Milan, Italy.
Organizational Affiliation: