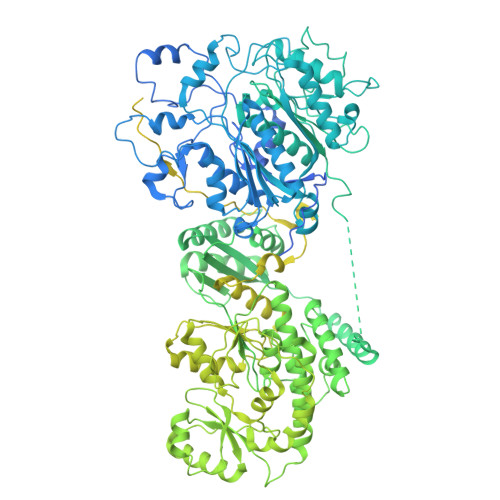
Cryo-electron microscopy structure of the di-domain core of Mycobacterium tuberculosis polyketide synthase 13, essential for mycobacterial mycolic acid synthesis.
Johnston, H.E., Batt, S.M., Brown, A.K., Savva, C.G., Besra, G.S., Futterer, K.(2024) Microbiology (Reading) 170
- PubMed: 39412527
- DOI: https://doi.org/10.1099/mic.0.001505
- Primary Citation of Related Structures:
9F48 - PubMed Abstract:
Mycobacteria are known for their complex cell wall, which comprises layers of peptidoglycan, polysaccharides and unusual fatty acids known as mycolic acids that form their unique outer membrane. Polyketide synthase 13 (Pks13) of Mycobacterium tuberculosis , the bacterial organism causing tuberculosis, catalyses the last step of mycolic acid synthesis prior to export to and assembly in the cell wall. Due to its essentiality, Pks13 is a target for several novel anti-tubercular inhibitors, but its 3D structure and catalytic reaction mechanism remain to be fully elucidated. Here, we report the molecular structure of the catalytic core domains of M. tuberculosis Pks13 (Mt-Pks13), determined by transmission cryo-electron microscopy (cryoEM) to a resolution of 3.4 Å. We observed a homodimeric assembly comprising the ketoacyl synthase (KS) domain at the centre, mediating dimerization, and the acyltransferase (AT) domains protruding in opposite directions from the central KS domain dimer. In addition to the KS-AT di-domains, the cryoEM map includes features not covered by the di-domain structural model that we predicted to contain a dimeric domain similar to dehydratases, yet likely lacking catalytic function. Analytical ultracentrifugation data indicate a pH-dependent equilibrium between monomeric and dimeric assembly states, while comparison with the previously determined structures of M. smegmatis Pks13 indicates architectural flexibility. Combining the experimentally determined structure with modelling in AlphaFold2 suggests a structural scaffold with a relatively stable dimeric core, which combines with considerable conformational flexibility to facilitate the successive steps of the Claisen-type condensation reaction catalysed by Pks13.
- School of Biosciences and Institute of Microbiology and Infection, University of Birmingham, Birmingham, B15 2TT, UK.
Organizational Affiliation: