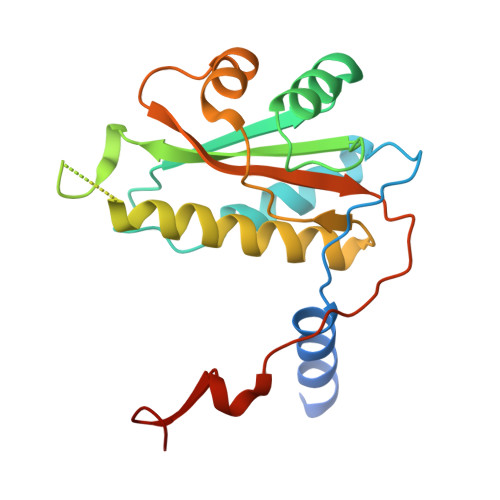
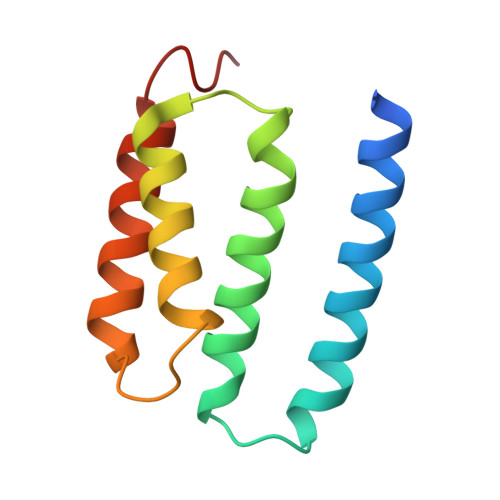
Broad substrate scope C-C oxidation in cyclodipeptides catalysed by a flavin-dependent filament.
Sutherland, E., Harding, C.J., du Monceau de Bergendal, T., Florence, G.J., Ackermann, K., Bode, B.E., Synowsky, S., Sundaramoorthy, R., Czekster, C.M.(2025) Nat Commun 16: 995-995
- PubMed: 39856061
- DOI: https://doi.org/10.1038/s41467-025-56127-y
- Primary Citation of Related Structures:
9EXV - PubMed Abstract:
Cyclic dipeptides are produced by organisms across all domains of life, with many exhibiting anticancer and antimicrobial properties. Oxidations are often key to their biological activities, particularly C-C bond oxidation catalysed by tailoring enzymes including cyclodipeptide oxidases. These flavin-dependent enzymes are underexplored due to their intricate three-dimensional arrangement involving multiple copies of two distinct small subunits, and mechanistic details underlying substrate selection and catalysis are lacking. Here, we determined the structure and mechanism of the cyclodipeptide oxidase from the halophile Nocardiopsis dassonvillei (NdasCDO), a component of the biosynthetic pathway for nocazine natural products. We demonstrated that NdasCDO forms filaments in solution, with a covalently bound flavin mononucleotide (FMN) cofactor at the interface between three distinct subunits. The enzyme exhibits promiscuity, processing various cyclic dipeptides as substrates in a distributive manner. The reaction is optimal at high pH and involves the formation of a radical intermediate. Pre-steady-state kinetics, a significant solvent kinetic isotope effect, and the absence of viscosity effects suggested that a step linked to FMN regeneration controlled the reaction rate. Our work elucidates the complex mechanistic and structural characteristics of this dehydrogenation reaction, positioning NdasCDO as a promising biocatalyst and expanding the FMN-dependent oxidase family to include enzyme filaments.
- University of St Andrews, School of Biology, North Haugh, Biomolecular Sciences Building, St Andrews, UK.
Organizational Affiliation: