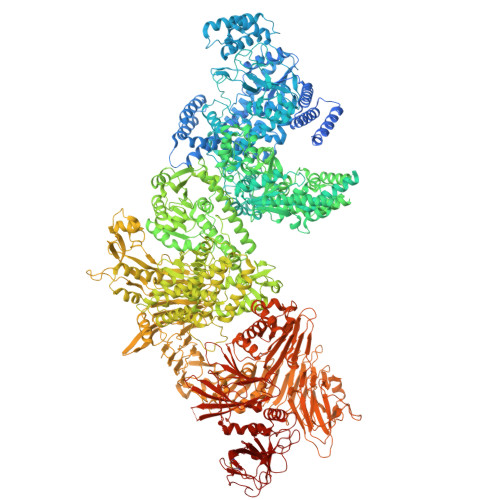
Structure of lymphostatin, a large multi-functional virulence factor of pathogenic Escherichia coli.
Griessmann, M., Rasmussen, T., Flegler, V.J., Kraft, C., Schneider, R., Hateley, M., Spantzel, L., Stevens, M.P., Borsch, M., Bottcher, B.(2025) Nat Commun 16: 5389-5389
- PubMed: 40562750
- DOI: https://doi.org/10.1038/s41467-025-60995-9
- Primary Citation of Related Structures:
9EUV, 9EUW, 9QB8, 9QBB, 9QHH - PubMed Abstract:
Lymphostatin is a key virulence factor of enteropathogenic and enterohaemorrhagic Escherichia coli, playing roles in bacterial colonisation of the gut and in the inhibition of lymphocyte proliferation and proinflammatory responses. The protein's glycosyltransferase and cysteine protease motifs are required for activity against lymphocytes, but high-resolution structural information has proven elusive. Here, we describe the structure of lymphostatin from enteropathogenic E. coli O127:H6, determined by electron cryo-microscopy at different pH values. We observe three conformations of a highly complex molecule with two glycosyltransferase domains, one PaToxP-like protease domain, an ADP-ribosyltransferase domain, a vertex domain and a delivery domain. Long linkers hold these domains together and occlude the catalytic sites of the N-terminal glycosyltransferase and protease domains. Lymphostatin binds to bovine T-lymphocytes and HEK-293T cells, forming clusters at the plasma membrane that are internalized. With six distinct domains, lymphostatin can be regarded as a multitool of pathogenic Escherichia coli, enabling complex interactions with host cells.
- University of Würzburg, Rudolf Virchow Center and Biocenter, Würzburg, Germany.
Organizational Affiliation: