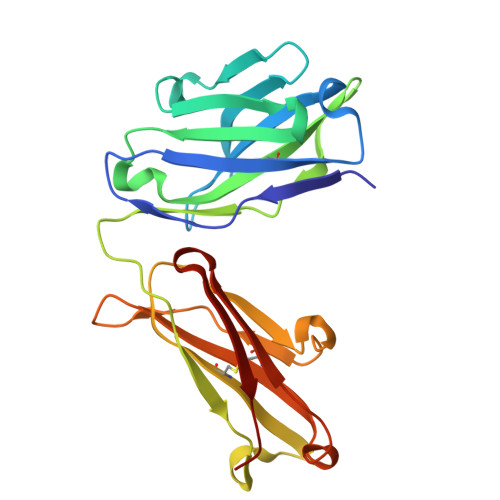
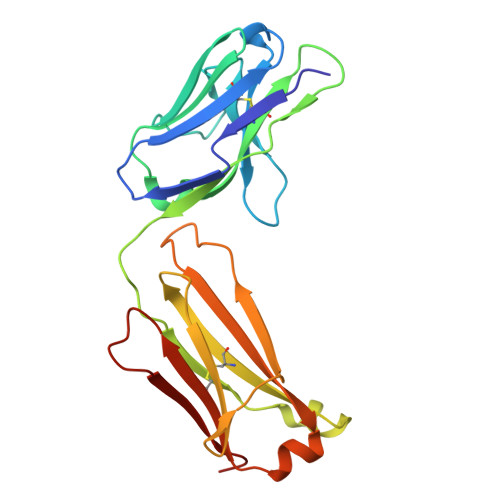
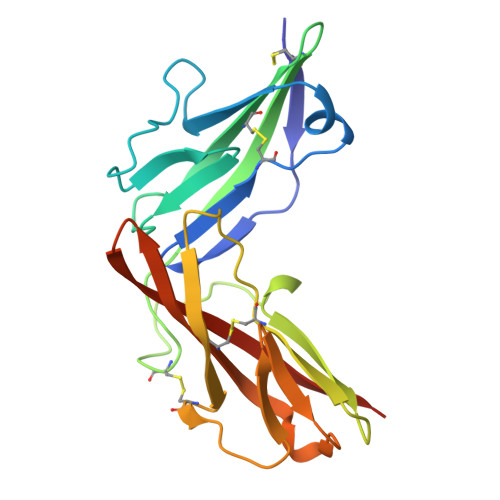
Discovery of selective low molecular weight interleukin-36 receptor antagonists by encoded library technologies.
Velcicky, J., Cremosnik, G., Scheufler, C., Meier, P., Wirth, E., Felber, R., Ramage, P., Schaefer, M., Kaiser, C., Lehmann, S., Kutil, R., Singeisen, S., Mueller-Ristig, D., Popp, S., Cebe, R., Lehr, P., Kaupmann, K., Erbel, P., Rohn, T.A., Giovannoni, J., Dumelin, C.E., Martiny-Baron, G.(2025) Nat Commun 16: 1669-1669
- PubMed: 39955284
- DOI: https://doi.org/10.1038/s41467-025-56601-7
- Primary Citation of Related Structures:
9ETH, 9ETI - PubMed Abstract:
Interleukin-36 receptor (IL-36R), belonging to the IL-1 receptor family, is crucial for host defense and tissue repair. Targeting cytokine receptors with low molecular weight (LMW) compounds remains challenging due to their interaction with the large surface area of cytokine. In this study, two encoded library technologies are used to identify LMW molecules binding to IL-36R's extracellular domain. The mRNA-based display technique identifies 36R-P138, a macrocyclic peptide blocking IL-36R signaling. Importantly, its optimized analog (36R-P192) also effectively suppresses expression of marker genes induced by IL-36 in human skin biopsies. DNA encoded libraries (DEL) screening delivers 36R-D481, a high affinity LMW IL-36R binder, effectively inhibiting IL-36 signaling. X-ray crystallography analysis reveals that both the cyclic peptide and DEL-compound bind to the IL-36R's D1 domain, potentially disrupting IL-36 cytokine binding. This study demonstrates that it is possible to target a cytokine receptor within the IL-1 receptor family using a small molecule ( < 1000 Da).
- Novartis Biomedical Research, Novartis Campus, CH-4002, Basel, Switzerland. juraj.velcicky@novartis.com.
Organizational Affiliation: