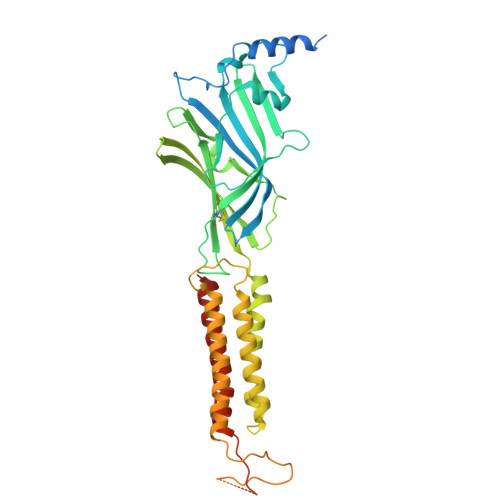
A brain-to-gut signal controls intestinal fat absorption.
Lyu, Q., Xue, W., Liu, R., Ma, Q., Kasaragod, V.B., Sun, S., Li, Q., Chen, Y., Yuan, M., Yang, Y., Zhang, B., Nie, A., Jia, S., Shen, C., Gao, P., Rong, W., Yu, C., Bi, Y., Zhang, C., Nan, F., Ning, G., Rao, Z., Yang, X., Wang, J., Wang, W.(2024) Nature 634: 936-943
- PubMed: 39261733
- DOI: https://doi.org/10.1038/s41586-024-07929-5
- Primary Citation of Related Structures:
9EQG - PubMed Abstract:
Although fat is a crucial source of energy in diets, excessive intake leads to obesity. Fat absorption in the gut is prevailingly thought to occur organ-autonomously by diffusion 1-3 . Whether the process is controlled by the brain-to-gut axis, however, remains largely unknown. Here we demonstrate that the dorsal motor nucleus of vagus (DMV) plays a key part in this process. Inactivation of DMV neurons reduces intestinal fat absorption and consequently causes weight loss, whereas activation of the DMV increases fat absorption and weight gain. Notably, the inactivation of a subpopulation of DMV neurons that project to the jejunum shortens the length of microvilli, thereby reducing fat absorption. Moreover, we identify a natural compound, puerarin, that mimics the suppression of the DMV-vagus pathway, which in turn leads to reduced fat absorption. Photoaffinity chemical methods and cryogenic electron microscopy of the structure of a GABA A receptor-puerarin complex reveal that puerarin binds to an allosteric modulatory site. Notably, conditional Gabra1 knockout in the DMV largely abolishes puerarin-induced intestinal fat loss. In summary, we discover that suppression of the DMV-vagus-jejunum axis controls intestinal fat absorption by shortening the length of microvilli and illustrate the therapeutic potential of puerarin binding to GABRA1 in fat loss.
- Department of Endocrine and Metabolic Diseases, Shanghai Institute of Endocrine and Metabolic Diseases, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine (SJTUSM), Shanghai, China.
Organizational Affiliation: