Enzymatic photocatalysis mediated by a genetically encoded thiourea
Taylor, C.J., Hardy, F.J., Green, A.P.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CT1.5 | 242 | synthetic construct | Mutation(s): 0 | 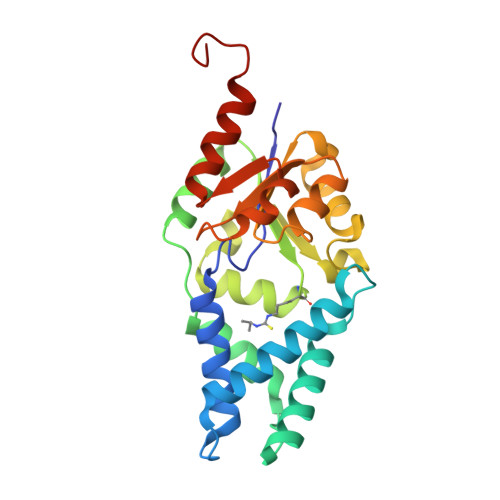 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MLA Query on MLA | B [auth A], J [auth A] | MALONIC ACID C3 H4 O4 OFOBLEOULBTSOW-UHFFFAOYSA-N |  | ||
| ACY Query on ACY | C [auth A], D [auth A] | ACETIC ACID C2 H4 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-N |  | ||
| NA Query on NA | E [auth A], F [auth A], G [auth A], H [auth A], I [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| A1H96 Query on A1H96 | A | L-PEPTIDE LINKING | C10 H21 N3 O2 S |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 69.919 | α = 90 |
| b = 69.919 | β = 90 |
| c = 122.243 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| DIALS | data reduction |
| DIALS | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Engineering and Physical Sciences Research Council | United Kingdom | EP/W524347/1 |
| Human Frontier Science Program (HFSP) | France | RGP0004/2022 |
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/M027023/1 |