Structural basis for cohesin release and control of genome looping by WAPL
Graham, J.J., Panne, D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cohesin subunit SA-2 | 981 | Homo sapiens | Mutation(s): 0 Gene Names: STAG2, SA2 | 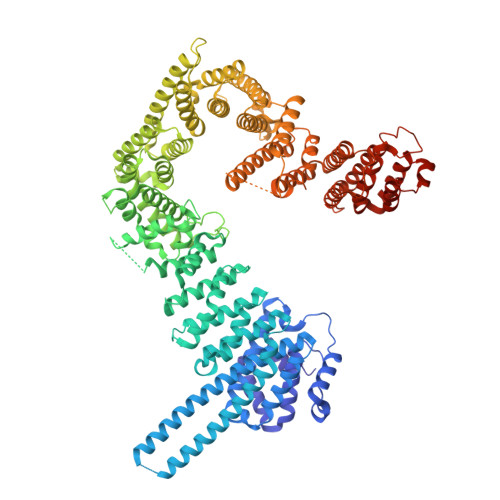 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q8N3U4 (Homo sapiens) Explore Q8N3U4 Go to UniProtKB: Q8N3U4 | |||||
PHAROS: Q8N3U4 GTEx: ENSG00000101972 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8N3U4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Double-strand-break repair protein rad21 homolog | 140 | Homo sapiens | Mutation(s): 0 Gene Names: RAD21, HR21, KIAA0078, NXP1, SCC1 | 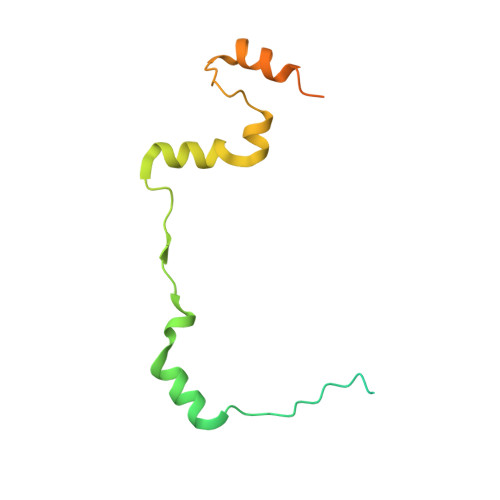 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O60216 (Homo sapiens) Explore O60216 Go to UniProtKB: O60216 | |||||
PHAROS: O60216 GTEx: ENSG00000164754 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O60216 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Wings apart-like protein homolog | 22 | Homo sapiens | Mutation(s): 0 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q7Z5K2 (Homo sapiens) Explore Q7Z5K2 Go to UniProtKB: Q7Z5K2 | |||||
PHAROS: Q7Z5K2 GTEx: ENSG00000062650 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7Z5K2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 78.433 | α = 90 |
| b = 107.44 | β = 90 |
| c = 181.913 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | 221881/Z/20/Z |
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MR/W001667/1 |