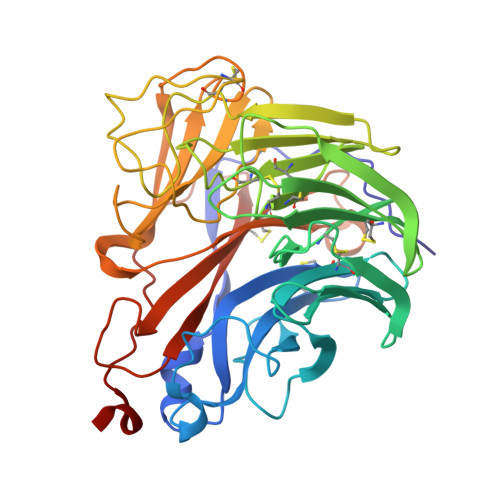
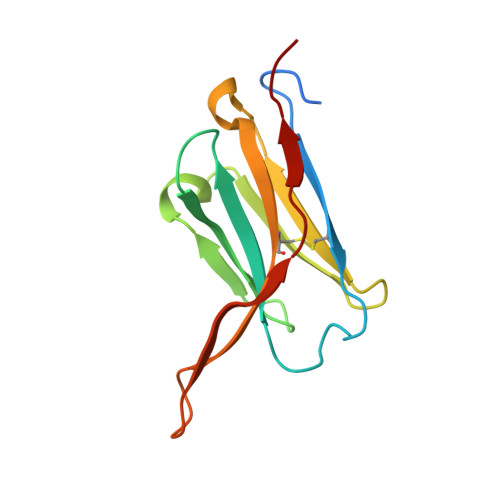
Structural convergence and water-mediated substrate mimicry enable broad neuraminidase inhibition by human antibodies.
Lederhofer, J., Borst, A.J., Nguyen, L., Gillespie, R.A., Williams, C.J., Walker, E.L., Raab, J.E., Yap, C., Ellis, D., Creanga, A., Tan, H.X., Do, T.H.T., Ravichandran, M., McDermott, A.B., Le Sage, V., Andrews, S.F., Graham, B.S., Wheatley, A.K., Reed, D.S., King, N.P., Kanekiyo, M.(2025) Nat Commun 16: 7068-7068
- PubMed: 40750617
- DOI: https://doi.org/10.1038/s41467-025-62339-z
- Primary Citation of Related Structures:
9EIT, 9EJE, 9EJF, 9O9V - PubMed Abstract:
Influenza has been responsible for multiple global pandemics and seasonal epidemics and claimed millions of lives. The imminent threat of a panzootic outbreak of avian influenza H5N1 virus underscores the urgent need for pandemic preparedness and effective countermeasures, including monoclonal antibodies (mAbs). Here, we characterize human mAbs that target the highly conserved catalytic site of viral neuraminidase (NA), termed NCS mAbs, and the molecular basis of their broad specificity. Cross-reactive NA-specific B cells were isolated by using stabilized NA probes of non-circulating subtypes. We found that NCS mAbs recognized multiple NAs of influenza A as well as influenza B NAs and conferred prophylactic protections in mice against H1N1, H5N1, and influenza B viruses. Cryo-electron microscopy structures of two NCS mAbs revealed that they rely on structural mimicry of sialic acid, the substrate of NA, by coordinating not only amino acid side chains but also water molecules, enabling inhibition of NA activity across multiple influenza A and B viruses, including avian influenza clade 2.3.4.4b H5N1 viruses. Our results provide a molecular basis for the broad reactivity and inhibitory activity of NCS mAbs targeting the catalytic site of NA through substrate mimicry.
- Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, 20892, USA.
Organizational Affiliation: