Distinct horizontal transfer mechanisms for type I and type V CRISPR-associated transposons.
Hu, K., Chou, C.W., Wilke, C.O., Finkelstein, I.J.(2024) Nat Commun 15: 6653-6653
- PubMed: 39103341
- DOI: https://doi.org/10.1038/s41467-024-50816-w
- Primary Citation of Related Structures:
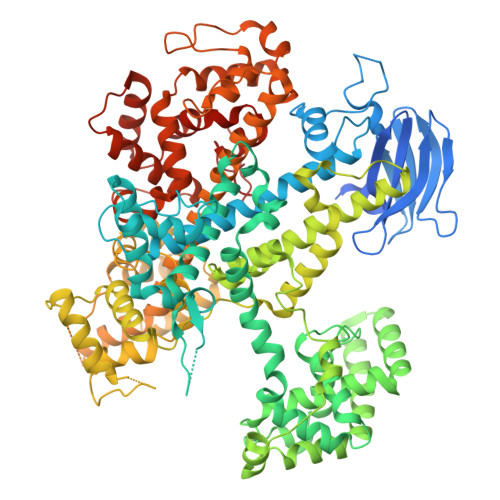
9EC8 - PubMed Abstract:
CASTs use both CRISPR-associated proteins and Tn7-family transposons for RNA-guided vertical and horizontal transmission. CASTs encode minimal CRISPR arrays but can't acquire new spacers. Here, we report that CASTs can co-opt defense-associated CRISPR arrays for horizontal transmission. A bioinformatic analysis shows that CASTs co-occur with defense-associated CRISPR systems, with the highest prevalence for type I-B and type V CAST sub-types. Using an E. coli quantitative transposition assay and in vitro reconstitution, we show that CASTs can use CRISPR RNAs from these defense systems. A high-resolution structure of the type I-F CAST-Cascade in complex with a type III-B CRISPR RNA reveals that Cas6 recognizes direct repeats via sequence-independent π - π interactions. In addition to using heterologous CRISPR arrays, type V CASTs can also transpose via an unguided mechanism, even when the S15 co-factor is over-expressed. Over-expressing S15 and the trans-activating CRISPR RNA or a single guide RNA reduces, but does not abrogate, off-target integration for type V CASTs. Our findings suggest that some CASTs may exploit defense-associated CRISPR arrays and that this fact must be considered when porting CASTs to heterologous bacterial hosts. More broadly, this work will guide further efforts to engineer the activity and specificity of CASTs for gene editing applications.
- Department of Molecular Biosciences, University of Texas at Austin, Austin, TX, 78712, USA. kh36969@utexas.edu.
Organizational Affiliation: