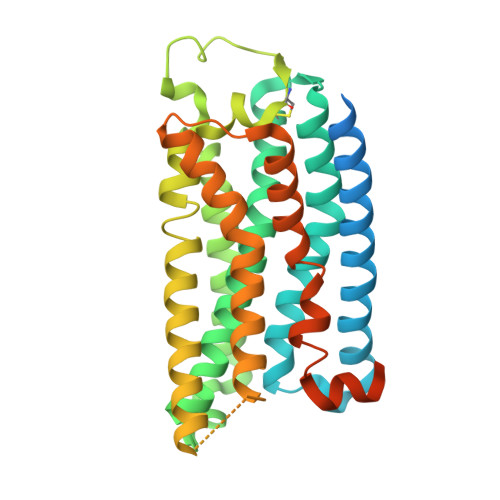
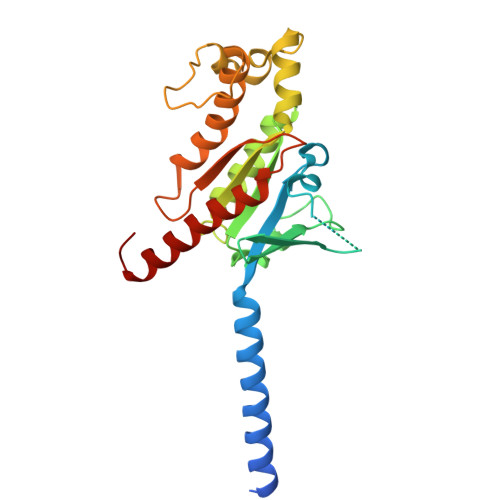
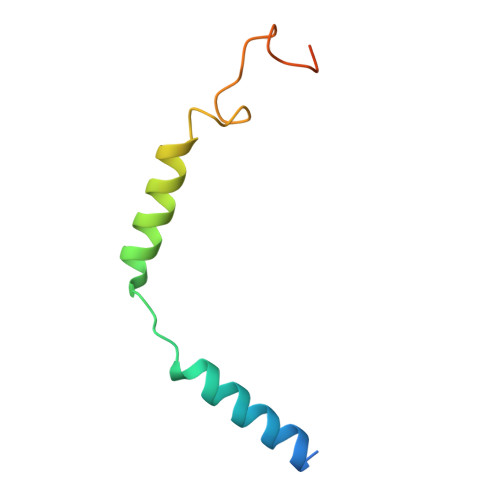
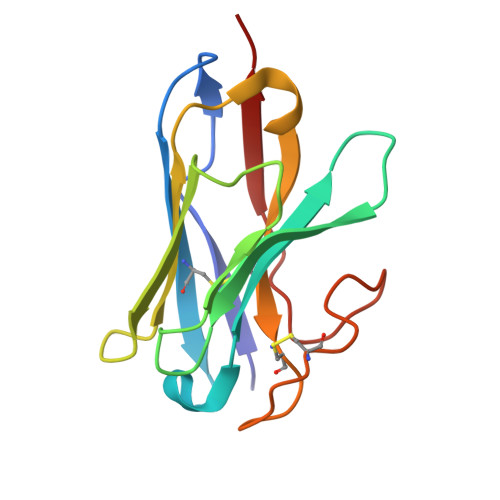
Molecular basis of ligand binding and receptor activation at the human A 3 adenosine receptor.
Zhang, L., Mobbs, J.I., Bennetts, F.M., Venugopal, H., Nguyen, A.T.N., Christopoulos, A., van der Es, D., Heitman, L.H., May, L.T., Glukhova, A., Thal, D.M.(2025) Nat Commun 16: 7674-7674
- PubMed: 40825947
- DOI: https://doi.org/10.1038/s41467-025-62872-x
- Primary Citation of Related Structures:
9EBH, 9EBI, 9EHS - PubMed Abstract:
Adenosine receptors (ARs: A 1 AR, A 2A AR, A 2B AR, and A 3 AR) are crucial therapeutic targets; however, developing selective, efficacious drugs for them remains a significant challenge. Here, we present high-resolution cryo-electron microscopy (cryo-EM) structures of the human A 3 AR in three distinct functional states: bound to the endogenous agonist adenosine, the clinically relevant agonist Piclidenoson, and the covalent antagonist LUF7602. These structures, complemented by mutagenesis and pharmacological studies, reveal an A 3 AR activation mechanism that involves an extensive hydrogen bond network from the extracellular surface down to the orthosteric binding site. In addition, we identify a cryptic pocket that accommodates the N 6 -iodobenzyl group of Piclidenoson through a ligand-dependent conformational change of M174 5.35 . Our comprehensive structural and functional characterisation of A 3 AR advances our understanding of adenosine receptor pharmacology and establishes a foundation for developing more selective therapeutics for various disorders, including inflammatory diseases, cancer, and glaucoma.
- Drug Discovery Biology, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, VIC, Australia.
Organizational Affiliation: