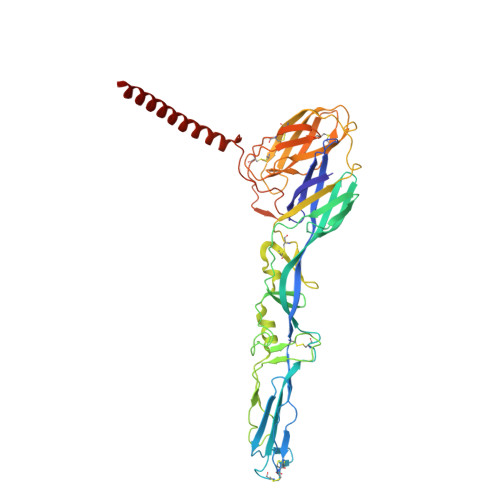
Structural basis for plasticity in receptor engagement by an encephalitic alphavirus.
Raju, S., Palakurty, S., Sariol, A., Wagoner, N., Adams, L.J., Hui, S., Klimstra, W.B., Fremont, D.H., Diamond, M.S.(2025) Cell 188: 2943
- PubMed: 40187344
- DOI: https://doi.org/10.1016/j.cell.2025.02.036
- Primary Citation of Related Structures:
9E96, 9E9Y, 9E9Z, 9EAU - PubMed Abstract:
The structural basis for shifts in receptor usage remains poorly understood despite the implications for virus adaptation and emergence. Western equine encephalitis virus (WEEV) strains exhibit different patterns of engagement for two of their entry receptors: very-low-density lipoprotein receptor (VLDLR) and protocadherin 10 (PCDH10). Using structural and functional studies, we show that while all WEEV strains have a lipoprotein class A (LA) domain binding site near the E1 fusion loop, VLDLR engagement requires a second binding site in E2 that can vary with single nucleotide substitutions. We also resolve a structure of PCDH10 bound to WEEV, which reveals interactions near the E1 fusion loop with residues that also mediate LA domain binding. Evolutionary analysis enabled the generation of a PCDH10 decoy that protects in vivo against all WEEV strains tested. Our experiments demonstrate how viruses can engage multiple receptors using shared determinants, which likely impacts cellular tropism and virulence.
- Department of Pathology & Immunology, Washington University School of Medicine, St. Louis, MO 63110, USA.
Organizational Affiliation: