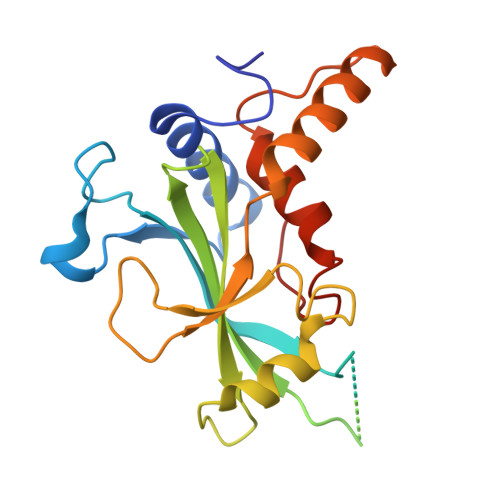
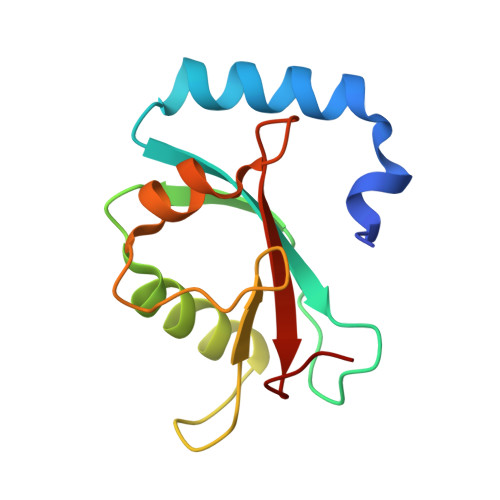
Structural Insights into the GABARAP-ATG3 Backside Interaction and Apo ATG3 Conformation.
Ohashi, K., Kroon, G.J., Otomo, T.(2025) Biochemistry 64: 3178-3189
- PubMed: 40628661
- DOI: https://doi.org/10.1021/acs.biochem.4c00485
- Primary Citation of Related Structures:
9E8P - PubMed Abstract:
Members of the ATG8 family of ubiquitin-like proteins (Ubls) are covalently attached to phosphatidylethanolamine (PE) on nascent autophagosomal membranes, where they recruit cargo receptors and promote membrane expansion. Although the overall lipidation pathway is well established, the molecular details-particularly those involving the E2 enzyme ATG3-remain incompletely defined. Here, we uncover a previously unrecognized, noncovalent binding mode between the mammalian ATG8 protein GABARAP and the backside of ATG3's catalytic E2 domain. In crystals, an isopeptide-linked GABARAP∼ATG3 conjugate self-assembles into a helical filament via this backside interface, mirroring architectures observed for canonical Ub/Ubl∼E2 conjugates. The E2 backside-binding surface on GABARAP is topologically distinct from those of other Ub/Ubl proteins and overlaps the LC3-interacting region (LIR) motif-binding site. Solution NMR confirms this interaction, and targeted mutagenesis shows that disrupting the interface impairs PE conjugation. Complementary NMR and AlphaFold modeling of apo ATG3 reveal an intramolecular contact between a segment of its flexible region (FR) and the catalytic core that suppresses conjugation. Together, these findings establish backside engagement as a critical feature of ATG8 lipidation and illuminate the dynamic architecture and regulation of ATG3.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Rd, La Jolla, California 92037, United States.
Organizational Affiliation: