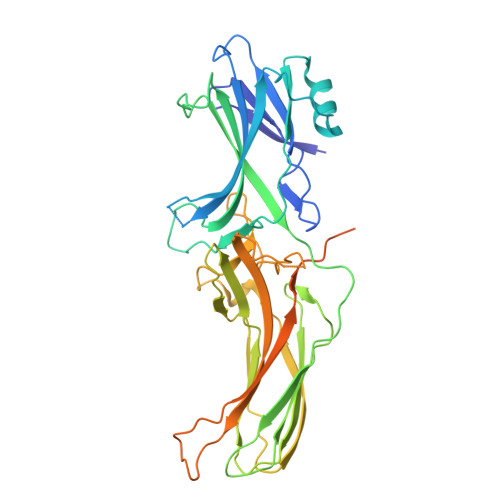
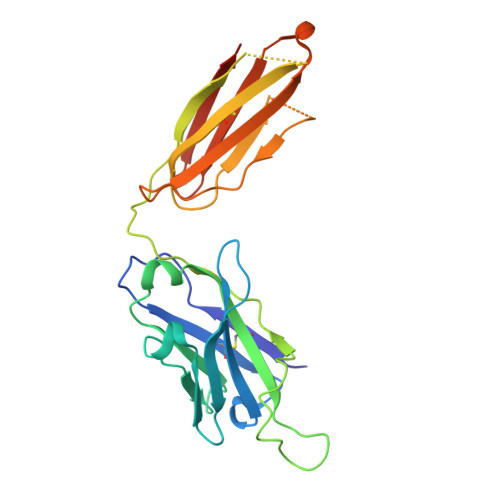
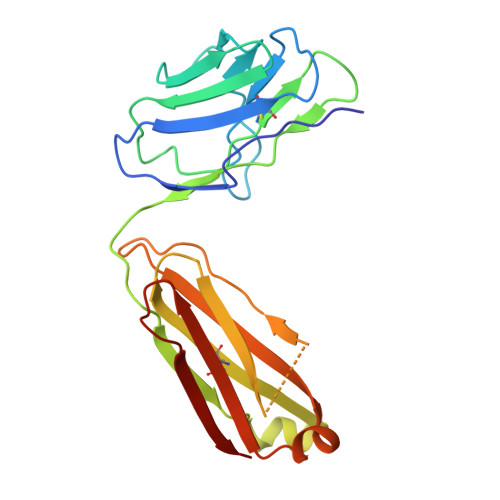
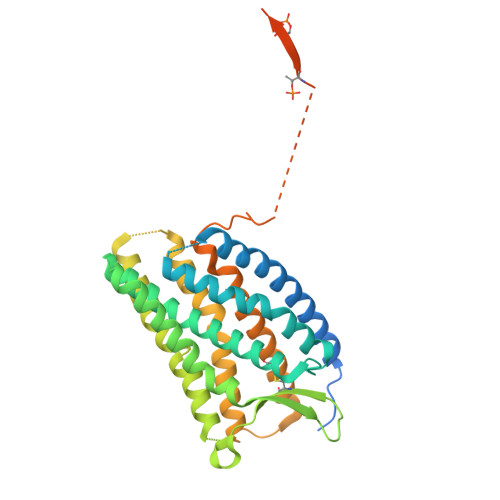
Effect of phosphorylation barcodes on arrestin binding to a chemokine receptor.
Chen, Q., Schafer, C.T., Mukherjee, S., Wang, K., Gustavsson, M., Fuller, J.R., Tepper, K., Lamme, T.D., Aydin, Y., Agrawal, P., Terashi, G., Yao, X.Q., Kihara, D., Kossiakoff, A.A., Handel, T.M., Tesmer, J.J.G.(2025) Nature 643: 280-287
- PubMed: 40399676
- DOI: https://doi.org/10.1038/s41586-025-09024-9
- Primary Citation of Related Structures:
8TII, 8TIL, 8TIN, 8TIO, 8VJ9, 9E82 - PubMed Abstract:
Unique phosphorylation 'barcodes' installed in different regions of an active seven-transmembrane receptor by different G-protein-coupled receptor (GPCR) kinases (GRKs) have been proposed to promote distinct cellular outcomes 1 , but it is unclear whether or how arrestins differentially engage these barcodes. Here, to address this, we developed an antigen-binding fragment (Fab7) that recognizes both active arrestin2 (β-arrestin1) and arrestin3 (β-arrestin2) without interacting with bound receptor polypeptides. We used Fab7 to determine the structures of both arrestins in complex with atypical chemokine receptor 3 (ACKR3) phosphorylated in different regions of its C-terminal tail by either GRK2 or GRK5 (ref. 2 ). The GRK2-phosphorylated ACKR3 resulted in more heterogeneous 'tail-mode' assemblies, whereas phosphorylation by GRK5 resulted in more rigid 'ACKR3-adjacent' assemblies. Unexpectedly, the finger loops of both arrestins engaged the micelle surface rather than the receptor intracellular pocket, with arrestin3 being more dynamic, partly because of its lack of a membrane-anchoring motif. Thus, both the region of the barcode and the arrestin isoform involved can alter the structure and dynamics of GPCR-arrestin complexes, providing a possible mechanistic basis for unique downstream cellular effects, such as the efficiency of chemokine scavenging and the robustness of arrestin binding in ACKR3.
- Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, IN, USA. qch2@iu.edu.
Organizational Affiliation: