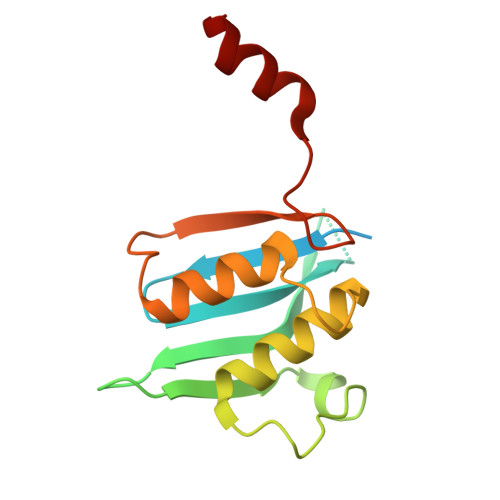
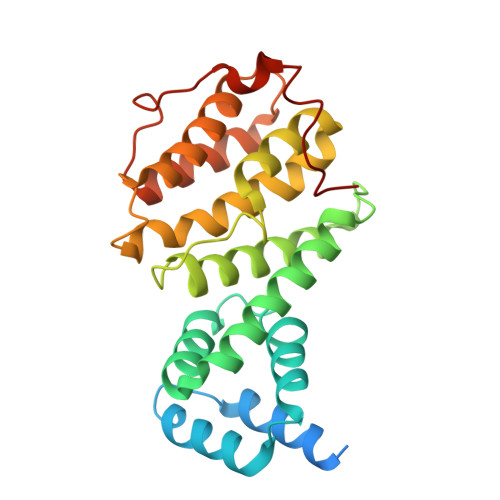
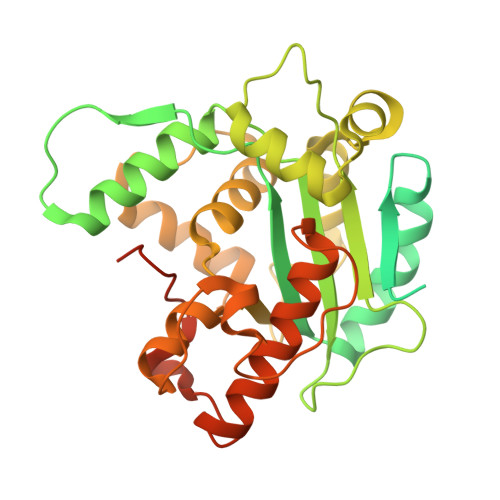
Intracellular pathogen effector reprograms host gene expression by inhibiting mRNA decay.
Levdansky, Y., Deme, J.C., Turner, D.J., Piczak, C.T., Pekovic, F., Valkov, A.L., Tarasov, S.G., Lea, S.M., Valkov, E.(2025) Nat Commun 16: 6452-6452
- PubMed: 40651939
- DOI: https://doi.org/10.1038/s41467-025-61194-2
- Primary Citation of Related Structures:
9E7T, 9E7U - PubMed Abstract:
Legionella pneumophila, an intracellular bacterial pathogen, injects effector proteins into host cells to manipulate cellular processes and promote its survival and proliferation. Here, we reveal a unique mechanism by which the Legionella effector PieF perturbs host mRNA decay by targeting the human CCR4-NOT deadenylase complex. High-resolution cryo-electron microscopy structures and biochemical analyses reveal that PieF binds with nanomolar affinity to the NOT7 and NOT8 catalytic subunits of CCR4-NOT, obstructing RNA access and displacing a catalytic Mg²⁺ ion from the active site. Additionally, PieF prevents NOT7/8 from associating with their partner deadenylases NOT6/6L, inhibiting the assembly of a functional deadenylase complex. Consequently, PieF robustly blocks mRNA poly(A) tail shortening and degradation with striking potency and selectivity for NOT7/8. This inhibition of deadenylation by PieF impedes cell cycle progression in human cells, revealing a novel bacterial strategy to modulate host gene expression.
- National Cancer Institute, National Institutes of Health, Frederick, MD, USA.
Organizational Affiliation: