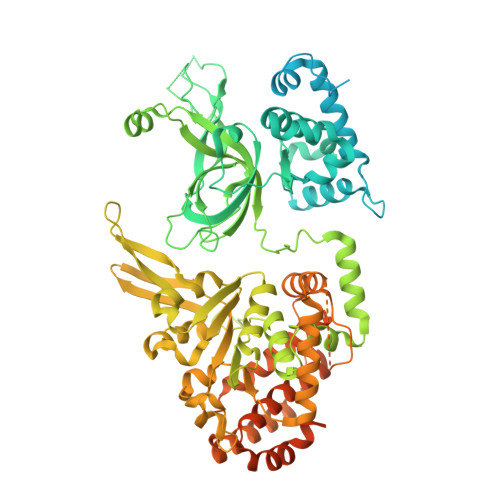
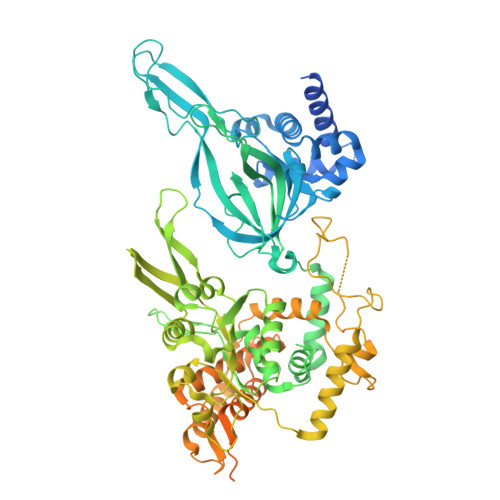
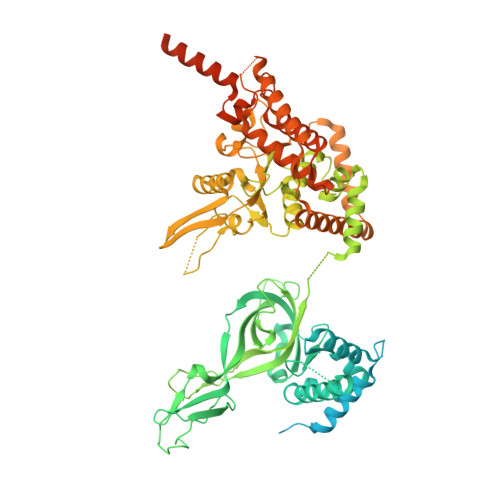
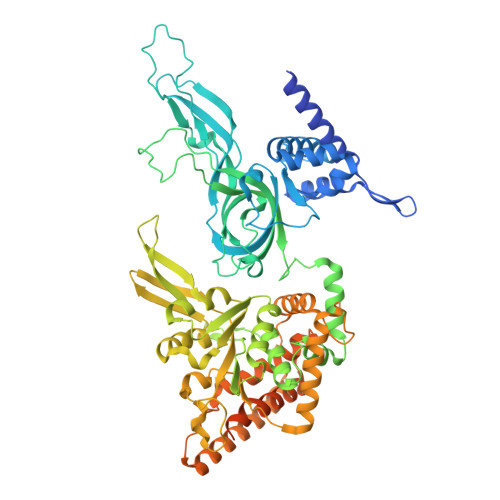
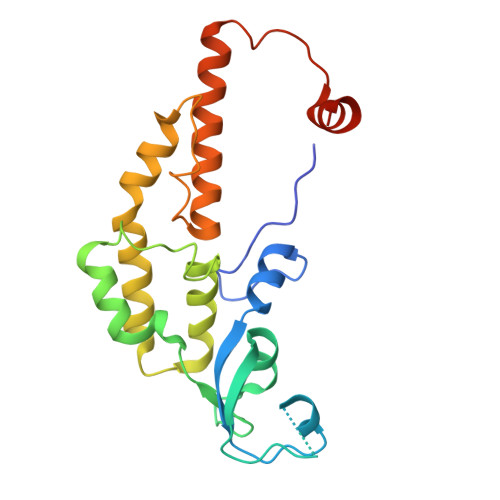
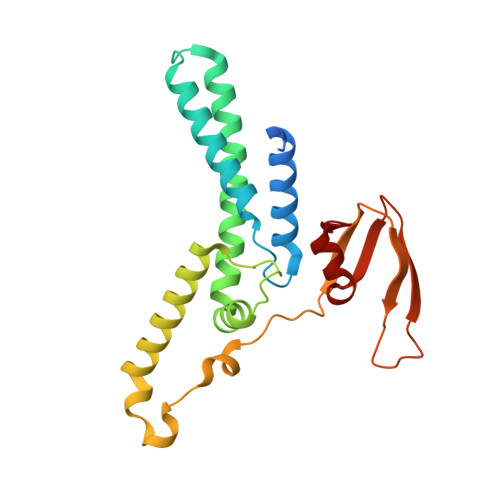
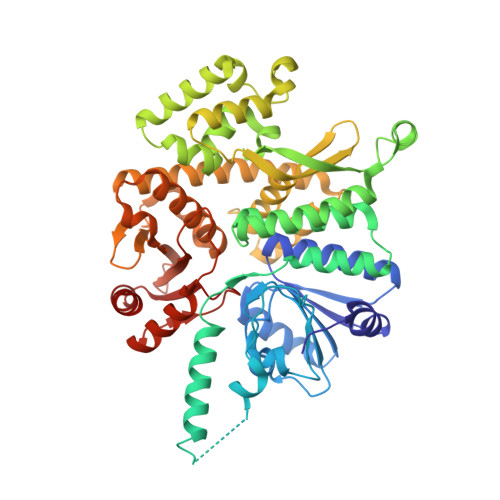
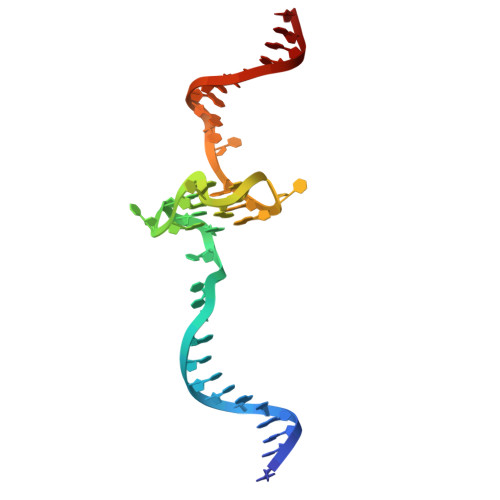
G-quadruplex-stalled eukaryotic replisome structure reveals helical inchworm DNA translocation.
Batra, S., Allwein, B., Kumar, C., Devbhandari, S., Bruning, J.G., Bahng, S., Lee, C.M., Marians, K.J., Hite, R.K., Remus, D.(2025) Science 387: eadt1978-eadt1978
- PubMed: 40048517
- DOI: https://doi.org/10.1126/science.adt1978
- Primary Citation of Related Structures:
9E2W, 9E2X, 9E2Y, 9E2Z - PubMed Abstract:
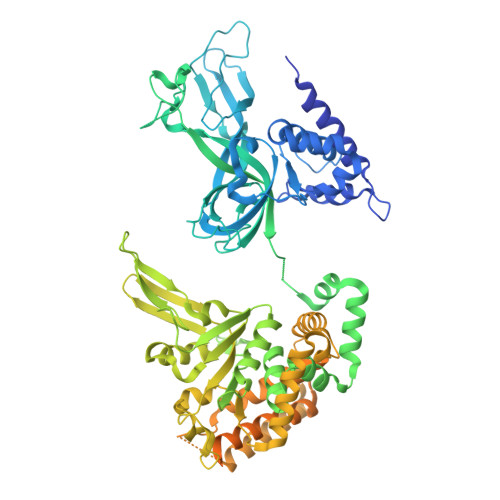
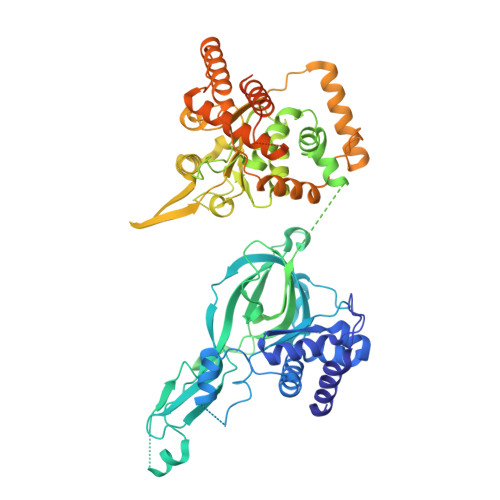
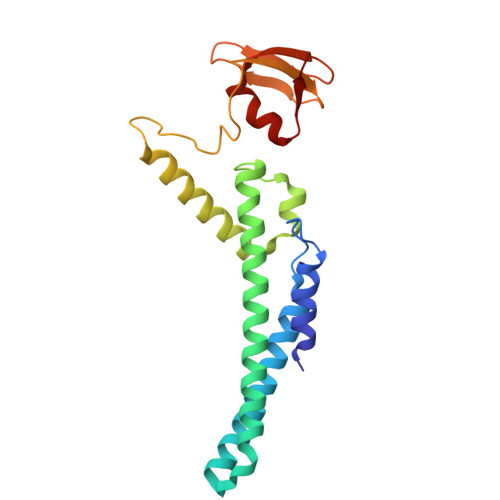
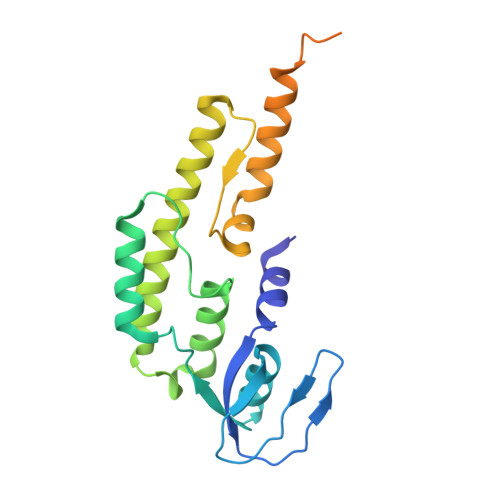
DNA G-quadruplexes (G4s) are non-B-form DNA secondary structures that threaten genome stability by impeding DNA replication. To elucidate how G4s induce replication fork arrest, we characterized fork collisions with preformed G4s in the parental DNA using reconstituted yeast and human replisomes. We demonstrate that a single G4 in the leading strand template is sufficient to stall replisomes by arresting the CMG helicase. Cryo-electron microscopy structures of stalled yeast and human CMG complexes reveal that the folded G4 is lodged inside the central CMG channel, arresting translocation. The G4 stabilizes the CMG at distinct translocation intermediates, suggesting an unprecedented helical inchworm mechanism for DNA translocation. These findings illuminate the eukaryotic replication fork mechanism under normal and perturbed conditions.
- Molecular Biology Program, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Organizational Affiliation: