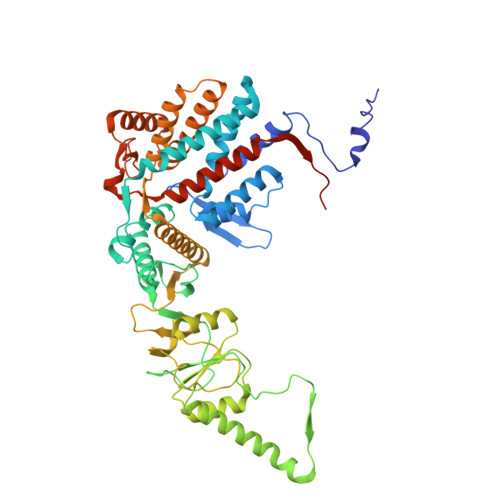
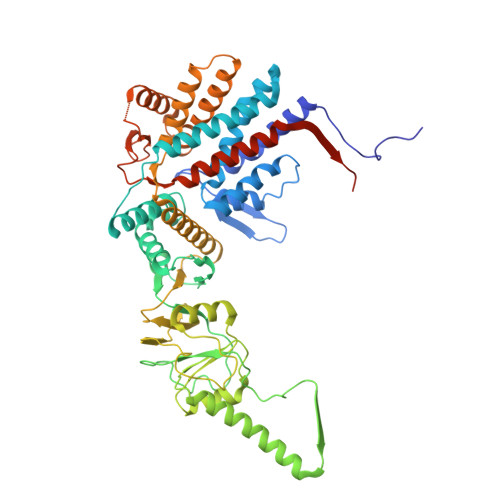
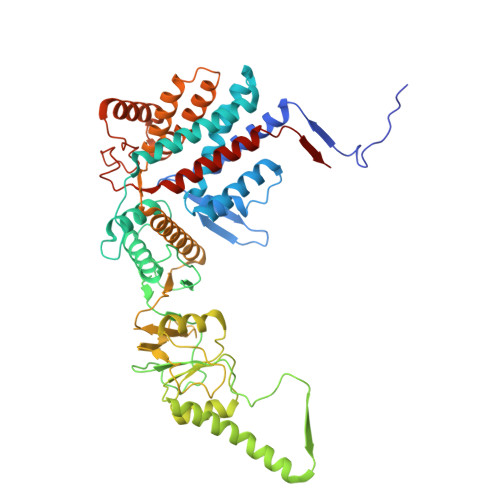
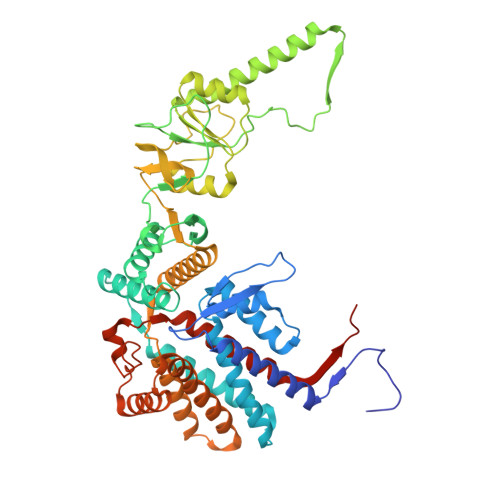
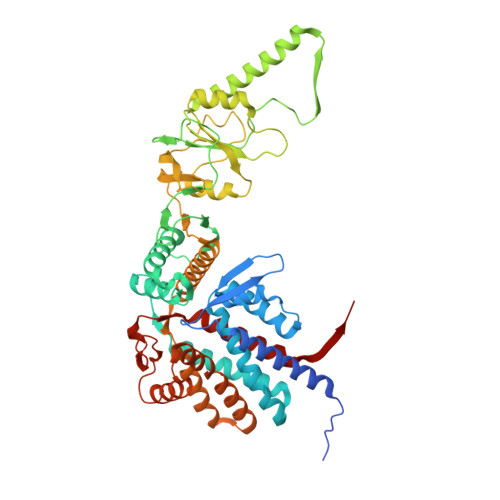
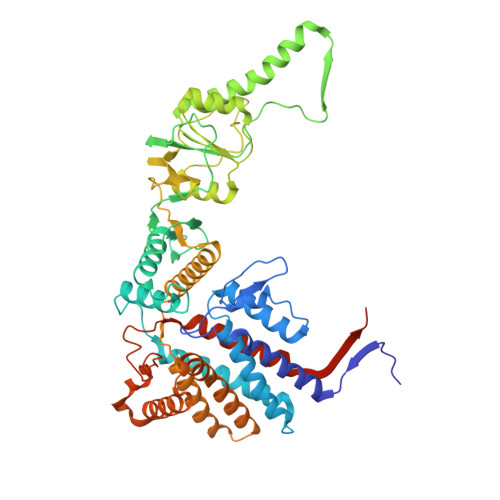
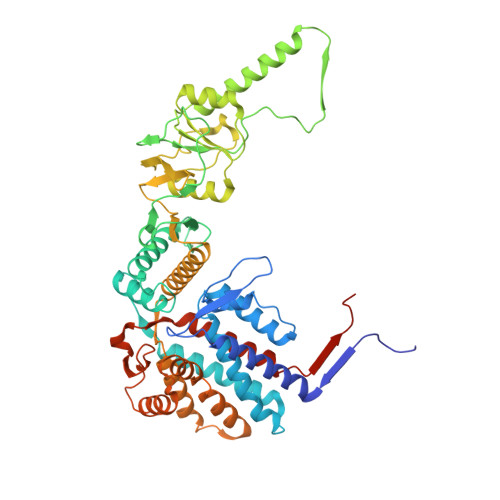
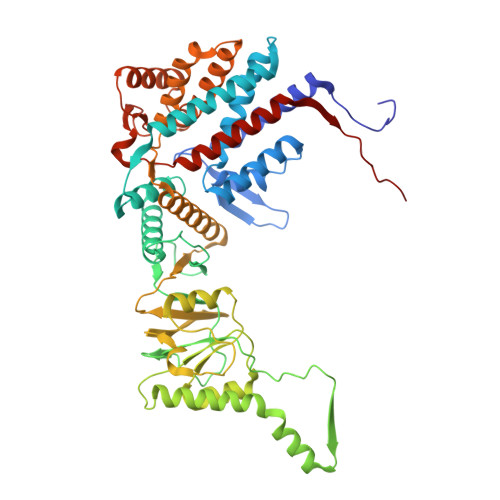
Visualizing dynamic tubulin folding in chaperonin TRiC from nonnative nucleus to final native state.
Zhao, Y., Schmid, M.F., Chiu, W.(2025) Nat Commun 16: 7878-7878
- PubMed: 40849413
- DOI: https://doi.org/10.1038/s41467-025-63016-x
- Primary Citation of Related Structures:
9E2S - PubMed Abstract:
The folding nucleus (FN) initiates and enables an efficient protein folding pathway. Despite its essential role, the FN has long remained cryptic. Here we directly visualize the tubulin FN consisting of a nonnative, partially assembled Rossmann fold, in the closed chamber of human chaperonin TRiC. Chaperonin TRiC interacts with nonnatively folded secondary structure elements of tubulin, stabilizing the nucleus poised for transition into its first native domain tertiary structure. Through progressive folding into the native state, we observe that the unfolded sequence of tubulin undergoes drastic spatial rearrangement in the TRiC chamber to sample the conformational space, mediated by the highly dynamic CCT tails. The observed presence of individual nonnative secondary structure elements first in the nonnative FN and then around the incrementally folded native domains supports the hypothesis that tubulin folding in TRiC is a hierarchical process of nucleation, condensation and propagation in cooperation with TRiC subunits.
- Department of Bioengineering, Stanford University, Stanford, CA, USA. yanyanz@stanford.edu.
Organizational Affiliation: