Structure of the twin-arginine protein translocation pathway core complex and the molecular basis for substrate recognition
Deme, J.C., Bryant, O.J., Berks, B.C., Lea, S.M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sec-independent protein translocase protein TatC | A [auth E], C [auth A], E [auth C] | 382 | Nitratifractor salsuginis | Mutation(s): 0 Gene Names: tatC, Nitsa_0634 | 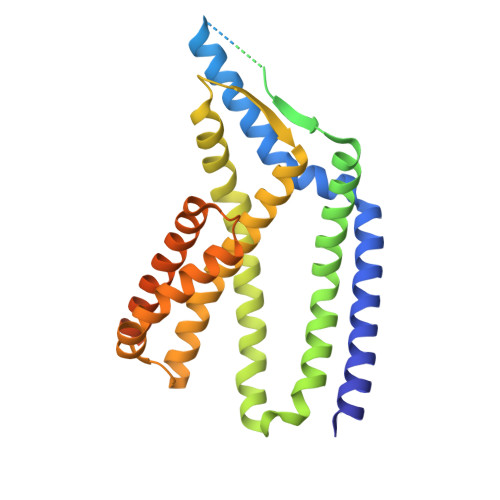 |
UniProt | |||||
Find proteins for E6X1G9 (Nitratifractor salsuginis (strain DSM 16511 / JCM 12458 / E9I37-1)) Explore E6X1G9 Go to UniProtKB: E6X1G9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | E6X1G9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sec-independent protein translocase protein TatB homolog | B [auth F], D [auth B], F [auth D] | 193 | Nitratifractor salsuginis | Mutation(s): 0 Gene Names: Nitsa_0635 | 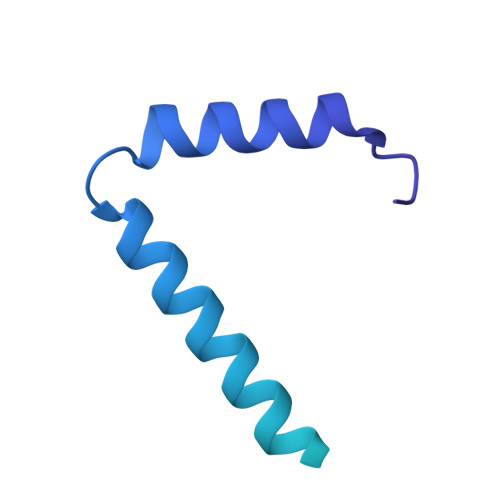 |
UniProt | |||||
Find proteins for E6X1H0 (Nitratifractor salsuginis (strain DSM 16511 / JCM 12458 / E9I37-1)) Explore E6X1H0 Go to UniProtKB: E6X1H0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | E6X1H0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MR/L000776/1 |
| Wellcome Trust | United Kingdom | 107929/Z/15/Z |
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | Intramural Research Program |