De novo design of D-peptide ligands: Application to influenza virus hemagglutinin.
Juraszek, J., Kadam, R.U., Branduardi, D., van Ameijde, J., Garg, D., Dailly, N., Jongeneelen, M., Vermond, J., Brakenhoff, J.P.J., Brandenburg, B., van Dongen, M.J.P., Vogels, R., Friesen, R.H.E., Wilson, I.A.(2025) Proc Natl Acad Sci U S A 122: e2426554122-e2426554122
- PubMed: 40577121
- DOI: https://doi.org/10.1073/pnas.2426554122
- Primary Citation of Related Structures:
9DXX - PubMed Abstract:
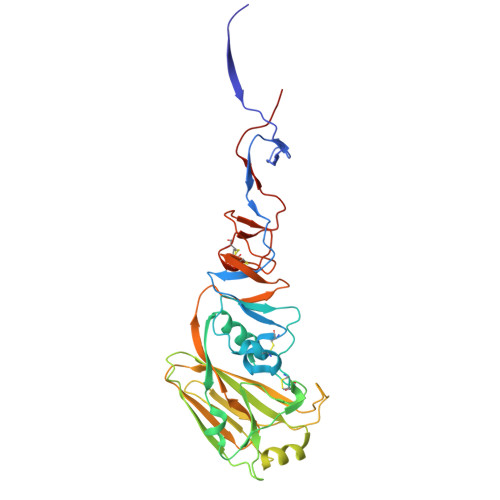
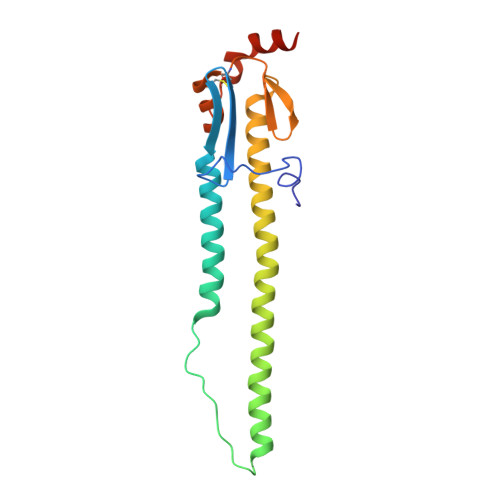
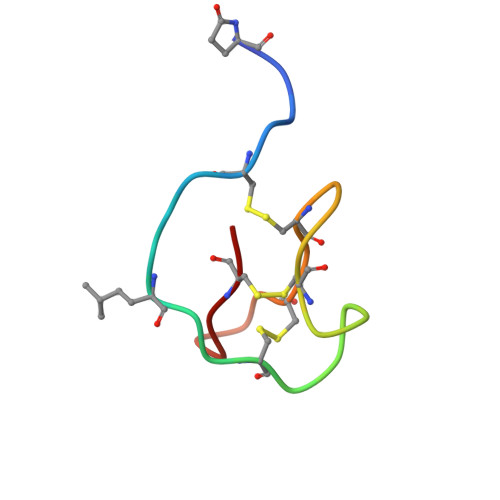
D-peptides hold great promise as therapeutics by alleviating the challenges of metabolic stability and immunogenicity in L-peptides. However, current D-peptide discovery methods are severely limited by specific size, structure, and the chemical synthesizability of their protein targets. Here, we describe a computational method for de novo design of D-peptides that bind to an epitope of interest on the target protein using Rosetta's hotspot-centric approach. The approach comprises identifying hotspot sidechains in a functional protein-protein interaction and grafting these side chains onto much smaller structured peptide scaffolds of opposite chirality. The approach enables more facile design of D-peptides and its applicability is demonstrated by design of D-peptidic binders of influenza A virus hemagglutinin, resulting in identification of multiple D-peptide lead series. The X-ray structure of one of the leads at 2.38 Å resolution verifies the validity of the approach. This method should be generally applicable to targets with detailed structural information, independent of molecular size, and accelerate development of stable, peptide-based therapeutics.
- Johnson and Johnson Innovative Medicine, Leiden 2333 CN, The Netherlands.
Organizational Affiliation: