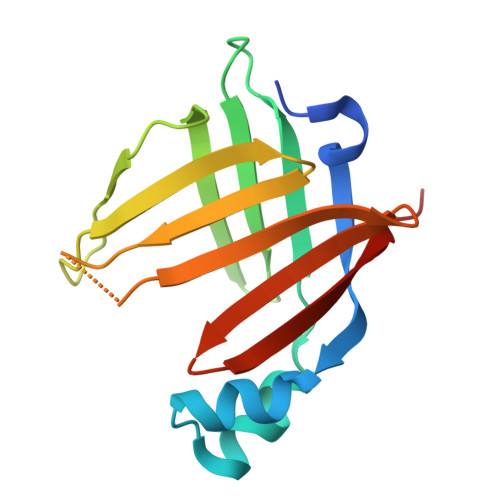
Structural dynamics and binding of Caenorhabditis elegans lifespan-extending lipid binding protein-3 to polyunsaturated fatty acids.
Cuevas, A.R., Tillman, M.C., Wang, M.C., Ortlund, E.A.(2025) Protein Sci 34: e5249-e5249
- PubMed: 39660930
- DOI: https://doi.org/10.1002/pro.5249
- Primary Citation of Related Structures:
9DXI - PubMed Abstract:
Intracellular lipid binding proteins (iLBPs) play crucial roles in lipid transport and cellular metabolism across the animal kingdom. Recently, a fat-to-neuron axis was described in Caenorhabditis elegans, in which lysosomal activity in the fat liberates polyunsaturated fatty acids (PUFAs) that signal to neurons and extend lifespan with durable fecundity. In this study, we investigate the structure and binding mechanisms of a lifespan-extending lipid chaperone, lipid binding protein-3 (LBP-3), which shuttles dihomo-γ-linolenic (DGLA) acid from intestinal fat to neurons. We present the first high-resolution crystal structure of LBP-3, which reveals a classic iLBP fold with an unexpected and unique homodimeric arrangement via interstrand interactions that is incompatible with ligand binding. We identify key ionic interactions that mediate DGLA binding within the lipid binding pocket. Molecular dynamics simulations further elucidate LBP-3's preferential binding to DGLA due to its rotational freedom and access to favorable binding conformations compared to other 20-carbon PUFAs. We also propose that LBP-3 dimerization may be a unique regulatory mechanism for lipid chaperones.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, Georgia, USA.
Organizational Affiliation: