Assembly and proteolytic activation human ClpXP defined by cryo-EM
Chen, W.C., Yang, J., Lander, G.C.To be published.
Experimental Data Snapshot
Starting Model: in silico
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial | 467 | Homo sapiens | Mutation(s): 0 Gene Names: CLPX EC: 3.6.4.10 | 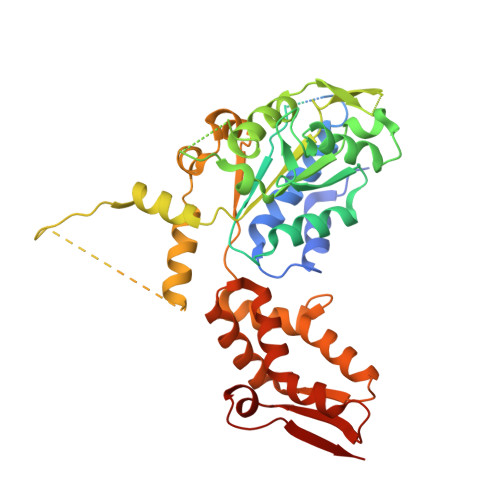 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O76031 (Homo sapiens) Explore O76031 Go to UniProtKB: O76031 | |||||
PHAROS: O76031 GTEx: ENSG00000166855 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O76031 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Unidentified endogenous substrate | 12 | Escherichia coli | Mutation(s): 0 |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ATP (Subject of Investigation/LOI) Query on ATP | I [auth A], K [auth B], M [auth C], O [auth D] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| ADP (Subject of Investigation/LOI) Query on ADP | P [auth E] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | H [auth A], J [auth B], L [auth C], N [auth D] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 4.3 |
| MODEL REFINEMENT | PHENIX | 1.21_5207 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (NSF, United States) | United States | -- |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | S10OD032467 |
| National Institutes of Health/National Institute of Neurological Disorders and Stroke (NIH/NINDS) | United States | NS095892 |