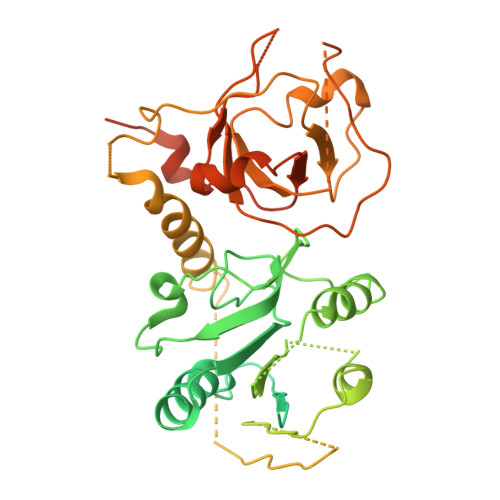
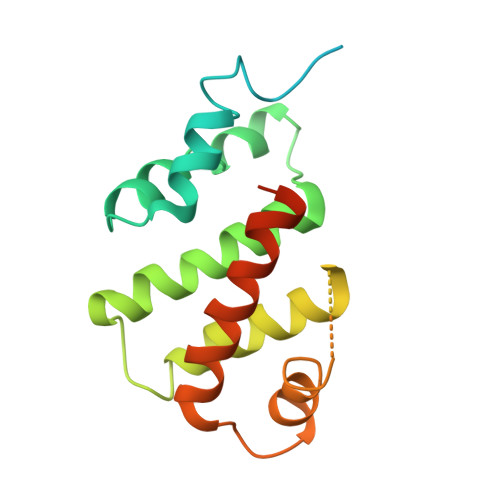
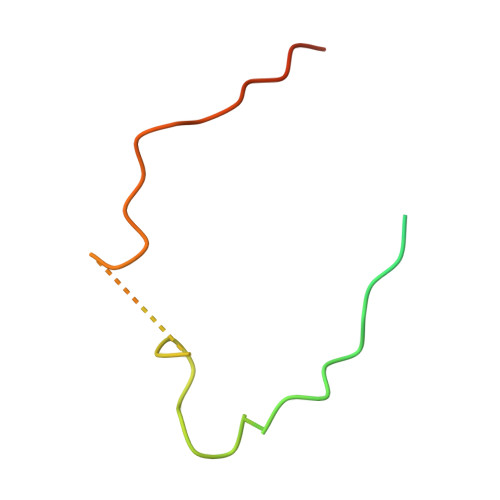
Molecular insights into the overall architecture of human rixosome.
Huang, J., Tong, L.(2025) Nat Commun 16: 3288-3288
- PubMed: 40195365
- DOI: https://doi.org/10.1038/s41467-025-58732-3
- Primary Citation of Related Structures:
9DUM, 9DUN, 9DUO - PubMed Abstract:
Rixosome is a conserved, multi-subunit protein complex that has critical roles in ribosome biogenesis and silencing of Polycomb target genes. The subunits of human rixosome include PELP1, WDR18, TEX10, LAS1L and NOL9, with LAS1L providing the endoribonuclease activity and NOL9 the RNA 5' kinase activity. We report here cryo-EM structures of the human PELP1-WDR18-TEX10 and LAS1L-NOL9 complexes and a lower-resolution model of the human PELP1-WDR18-LAS1L complex. The structures reveal the overall organization of the human rixosome core scaffold of PELP1-WDR18-TEX10-LAS1L and indicate how the LAS1L-NOL9 endonuclease/kinase catalytic module is recruited to this core scaffold. Each TEX10 molecule has two regions of contact with WDR18, while the helix at the C terminus of WDR18 interacts with the helical domain of LAS1L. The structural observations are supported by our mutagenesis studies. Mutations in both WDR18-TEX10 contact regions can block the binding of TEX10, while truncation of the C-terminal helix of WDR18 can abolish the binding of LAS1L. The structures also reveal substantial conformational differences for TEX10 between the PELP1-WDR18-TEX10 complex alone and that in complex with pre-ribosome.
- Department of Biological Sciences, Columbia University, New York, NY, USA.
Organizational Affiliation: