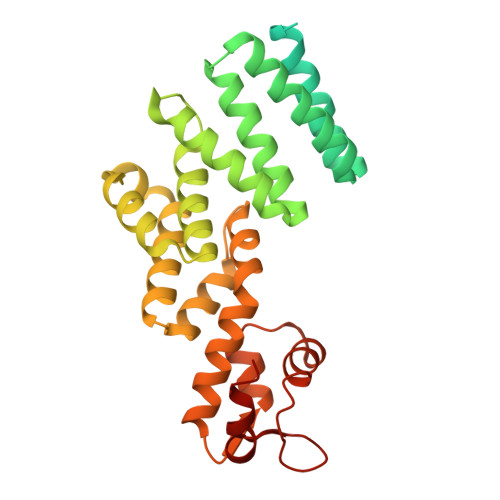
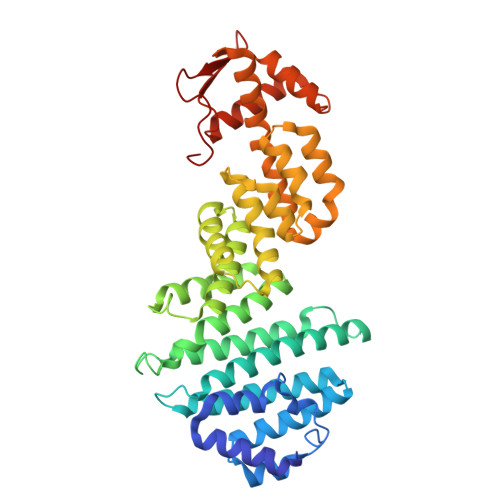
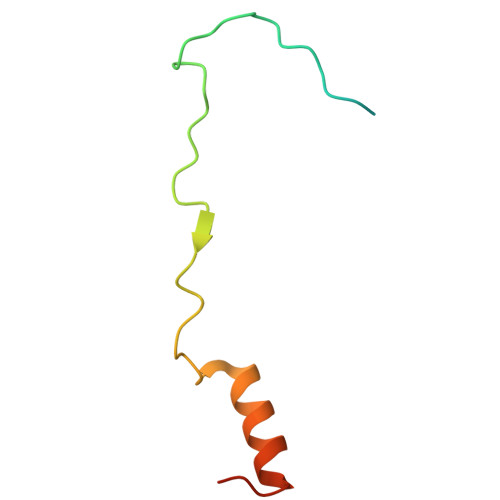
Structural mechanism of DDX39B regulation by human TREX-2 and a related complex in mRNP remodeling.
Clarke, B.P., Gao, S., Mei, M., Xie, D., Angelos, A.E., Vazhavilla, A., Hill, P.S., Cagatay, T., Batten, K., Shay, J.W., Xie, Y., Fontoura, B.M.A., Ren, Y.(2025) Nat Commun 16: 5471-5471
- PubMed: 40595470
- DOI: https://doi.org/10.1038/s41467-025-60547-1
- Primary Citation of Related Structures:
9DLP, 9DLR, 9DLV - PubMed Abstract:
Nuclear export of mRNAs in the form of messenger ribonucleoprotein particles (mRNPs) is an obligatory step for eukaryotic gene expression. The DEAD-box ATPase DDX39B (also known as UAP56) is a multifunctional regulator of nuclear mRNPs. How DDX39B mediates mRNP assembly and export in a controlled manner remains elusive. Here, we identify a novel complex TREX-2.1 localized in the nucleus that facilitates the release of DDX39B from the mRNP. TREX-2.1 is composed of three subunits, LENG8, PCID2, and DSS1, and shares the latter two subunits with the nuclear pore complex-associated TREX-2 complex. Cryo-EM structures of TREX-2.1/DDX39B and TREX-2/DDX39B identify a conserved trigger loop in the LENG8 and GANP subunit of the respective TREX-2.1 and TREX-2 complex that is critical for DDX39B regulation. RNA sequencing from LENG8 knockdown cells shows that LENG8 influences the nucleocytoplasmic ratio of a subset of mRNAs with high GC content. Together, our findings lead to a mechanistic understanding of the functional cycle of DDX39B and its regulation by TREX-2 and TREX-2.1 in mRNP processing.
- Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, TN, USA.
Organizational Affiliation: