Biochemical, structural, and biologial studies of Streptococcus pneumoniae GapN reveal a key metabolic player necessary for host infection.
Eunjeong, L., Elan, Z.E.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | 474 | Streptococcus pneumoniae | Mutation(s): 0 Gene Names: gapN, spr1028 EC: 1.2.1.9 | 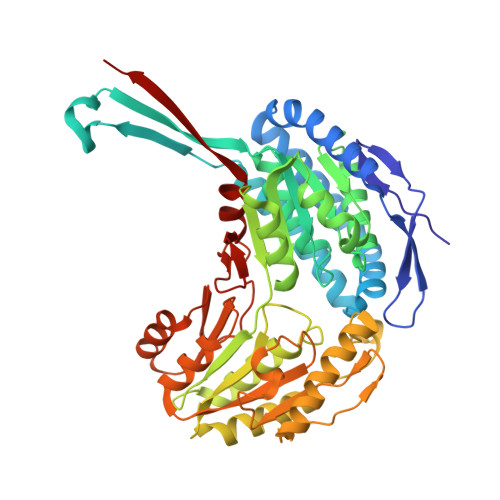 | |
UniProt | |||||
Find proteins for Q8DPS7 (Streptococcus pneumoniae (strain ATCC BAA-255 / R6)) Explore Q8DPS7 Go to UniProtKB: Q8DPS7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8DPS7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAP (Subject of Investigation/LOI) Query on NAP | E [auth A], F [auth B], G [auth C], H [auth D] | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE C21 H28 N7 O17 P3 XJLXINKUBYWONI-NNYOXOHSSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |