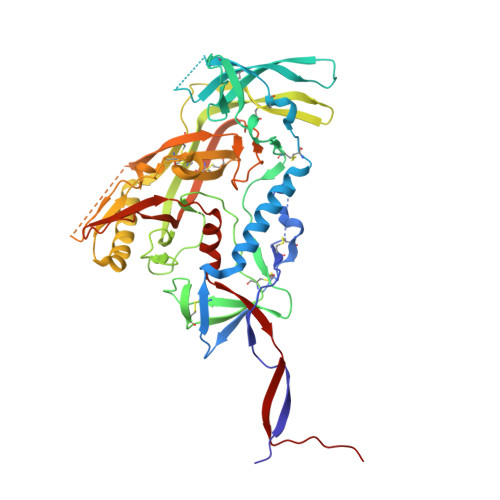
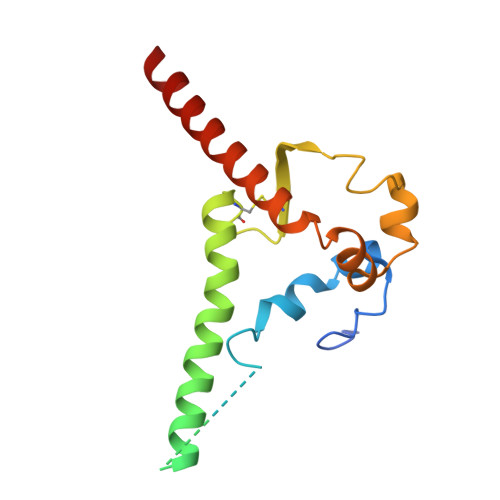
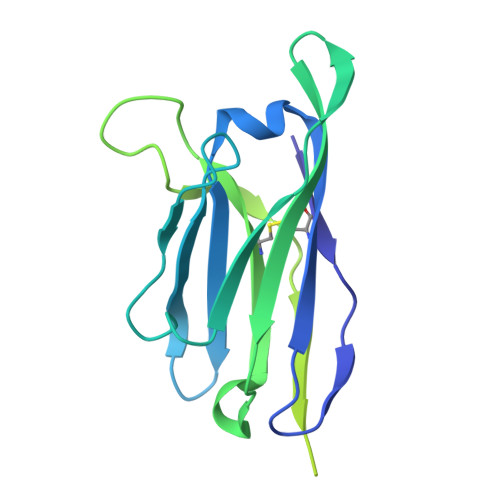
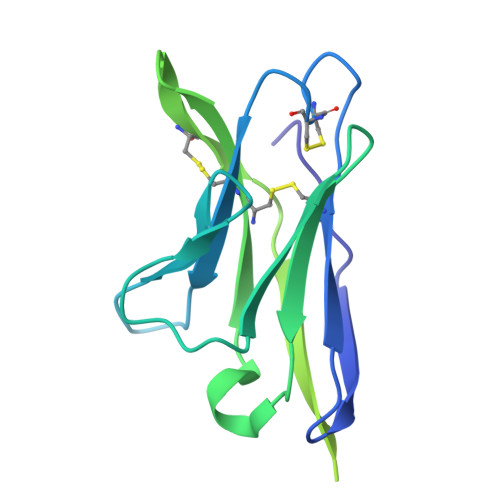
Deep Mining of the Human Antibody Repertoire Identifies Frequent and Genetically Diverse CDRH3 Topologies Targetable by Vaccination.
Habib, R., Solieva, S.O., Lin, Z.J., Ghosh, S., Bayruns, K., Singh, M., Agostino, C.J., Tursi, N.J., Sowers, K.J., Huang, J., Roark, R.S., Purwar, M., Park, Y., Ayyanathan, K., Li, H., Carey, J.W., Kim, A., Park, J., McCanna, M.E., Skelly, A.N., Chokkalingam, N., Kriete, S., Shupin, N., Huynh, A., Walker, S., Sadeesh, R., Laenger, N., Du, J., Cui, J., Patel, A., Escolano, A., Kwong, P.D., Shapiro, L., Bowman, G.R., Hahn, B.H., Shaw, G.M., Weiner, D.B., Pallesen, J., Kulp, D.W.(2025) bioRxiv
- PubMed: 41332666
- DOI: https://doi.org/10.1101/2024.10.04.616739
- Primary Citation of Related Structures:
9DHW, 9DIM - PubMed Abstract:
Germline targeting vaccination strategies against highly variable pathogens such as HIV aim to elicit broadly neutralizing antibodies (bnAbs) with particular immunogenetic or structural features. The V2 apex of the HIV Env protein is a promising target for a class of bnAbs that contain conserved structural motifs in the heavy chain complementarity determining region 3 (CDRH3). Here, we show that these structural motifs are targetable by vaccination by characterizing V2 apex 'axe-like' CDRH3s in the human repertoire and developing new immunogens capable of engaging them. We determined the frequency and diversity of axe-like CDHR3s in healthy human donors using a series of structural informatics approaches, finding these precursors in nearly 90% of donors. Axe-targeting immunogens based on the HIV Env Q23.17 bound axe-like precursors in cryo-EM structures, induced V2 apex-specific antibody responses in humanized mice, and induced axe-like heterologous neutralizing antibodies in rhesus macaques infected with a germline-targeted simian-human immunodeficiency virus. These results illustrate a new structure-guided immunoinformatic vaccine design paradigm that can be employed to elicit immunogenetically diverse yet structurally conserved classes of antibodies. Many broadly neutralizing antibodies (bnAbs) utilize modes of epitope recognition dominated by the antibody complementarity determining region 3 (CDRH3). The CDRH3 is the most diverse part of the antibody, posing a challenge for germline targeting vaccine designs that aim to elicit antibodies with particular immunogenetic features. Vaccine design strategies that accommodate CDRH3 variability are therefore needed. Many HIV Env V2 apex bnAbs share "axe-like" CDRH3 microfolds that arise from diverse immunogenetic origins. Here we determined the frequency in humans of B cells with such CDRH3 topologies and designed immunogens to engage their precursors. This work opens a path toward vaccines that engage specific structural classes of B cells, thereby advancing the rational design of immunogens for HIV and other pathogens.