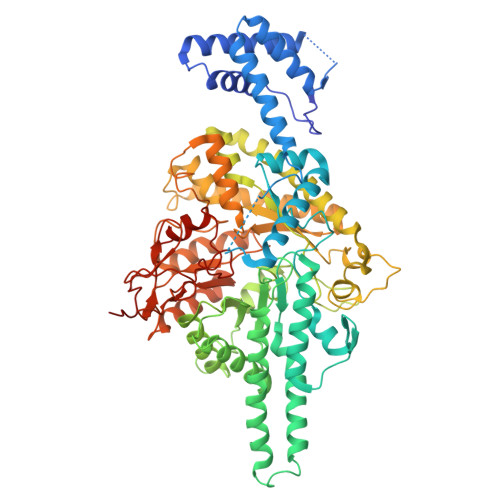
How ATP and dATP reposition class III ribonucleotide reductase cone domains to regulate enzyme activity.
Andree, G.A., Miller-Brown, K.R., Zhao, Z., Smith, A.K., Dawson, C.D., Deredge, D.J., Drennan, C.L.(2025) Sci Adv 11: eady9156-eady9156
- PubMed: 41313771
- DOI: https://doi.org/10.1126/sciadv.ady9156
- Primary Citation of Related Structures:
9D3X, 9DAU, 9DCA - PubMed Abstract:
Ribonucleotide reductases (RNRs) catalyze the conversion of ribonucleotides to deoxyribonucleotides. In the majority of cases, RNR activity is allosterically regulated by the cellular 2'-deoxyadenosine 5'-triphosphate (dATP)/adenosine 5'-triphosphate (ATP) ratio. To investigate allosteric activity regulation in anaerobic or class III (glycyl radical containing) RNRs, we determine cryo-electron microscopy structures of the class III RNR from Streptococcus thermophilus (StNrdD). We find that StNrdD's regulatory "cone" domains adopt markedly different conformations depending on whether the activator ATP or the inhibitor dATP is bound and that these different conformations alternatively position an "active site flap" toward the active site (ATP-bound) or away (dATP-bound). In contrast, the position of the glycyl radical domain is unaffected by the cone domain conformations, suggesting that StNrdD activity is regulated through control of substrate binding rather than control of radical transfer. Hydrogen-deuterium exchange mass spectrometry and mutagenesis support the structural findings. In addition, our structural data provide insight into the molecular basis by which ATP and dATP binding lead to the observed differential cone domain conformations.
- Department of Chemistry, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Organizational Affiliation: