Regulation of STING activation by phosphoinositide and cholesterol
Li, J., Tan, J.X., Chen, Z.J., Zhang, X., Bai, X.(2026) Nature
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2026) Nature
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Stimulator of interferon genes protein | 354 | Homo sapiens | Mutation(s): 0 Gene Names: STING1, ERIS, MITA, TMEM173 | 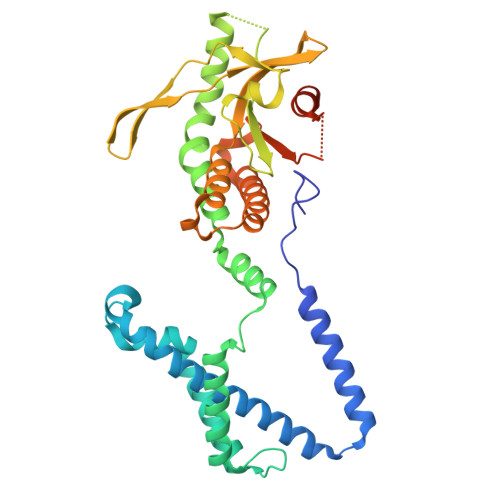 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q86WV6 (Homo sapiens) Explore Q86WV6 Go to UniProtKB: Q86WV6 | |||||
PHAROS: Q86WV6 GTEx: ENSG00000184584 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q86WV6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| A1BBH (Subject of Investigation/LOI) Query on A1BBH | E [auth A], J [auth C] | (2R)-3-{[(R)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl di[(9Z)-octadec-9-enoate] C45 H85 O19 P3 NCQJPYRCYOFNGI-BVBQJIMISA-N |  | ||
| 1SY (Subject of Investigation/LOI) Query on 1SY | G [auth B], I [auth C] | cGAMP C20 H24 N10 O13 P2 XRILCFTWUCUKJR-INFSMZHSSA-N |  | ||
| 9IM (Subject of Investigation/LOI) Query on 9IM | H [auth B], L [auth D] | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide C25 H19 Cl F4 N2 O2 OEBVYNULNKSONR-UHFFFAOYSA-N |  | ||
| Y01 (Subject of Investigation/LOI) Query on Y01 | F [auth A], K [auth C] | CHOLESTEROL HEMISUCCINATE C31 H50 O4 WLNARFZDISHUGS-MIXBDBMTSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Cancer Institute (NIH/NCI) | United States | R01CA273595 |
| Welch Foundation | United States | I-1702 |
| Welch Foundation | United States | I-1944 |