Structural basis of nucleosome recognition by the conserved Dsup and HMGN nucleosome-binding motif.
Alegrio-Louro, J., Cruz-Becerra, G., Kassavetis, G.A., Kadonaga, J.T., Leschziner, A.E.(2025) Genes Dev 39: 1155-1161
- PubMed: 40721296
- DOI: https://doi.org/10.1101/gad.352720.125
- Primary Citation of Related Structures:
9D3K, 9D3L, 9D3M, 9D3N, 9D3O, 9D3P, 9D3Q, 9D3R, 9D3S, 9D3T - PubMed Abstract:
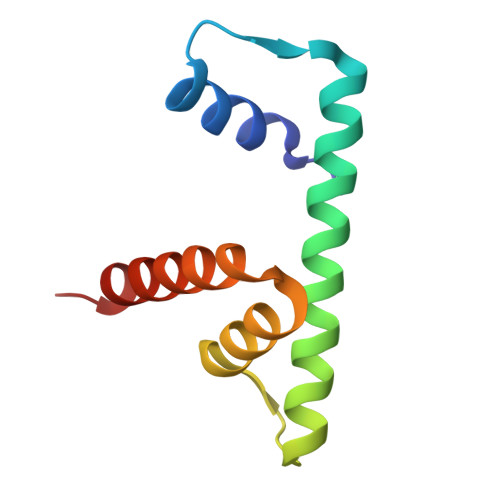
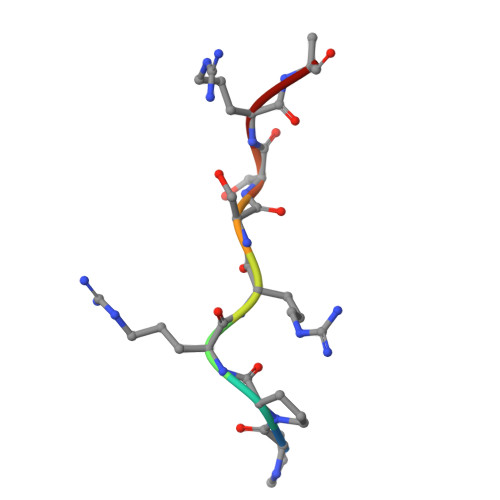
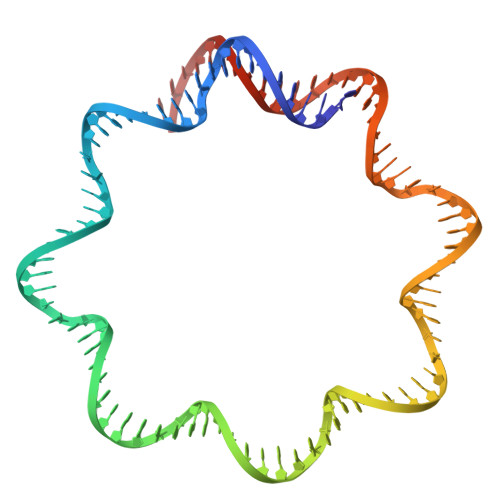
The tardigrade damage suppressor (Dsup) and vertebrate high-mobility group N (HMGN) proteins bind specifically to nucleosomes via a conserved motif whose structure has not been experimentally determined. Here we used cryo-EM to show that both proteins bind to the nucleosome acidic patch via analogous arginine anchors with one molecule bound to each face of the nucleosome. We additionally used the natural promoter-containing 5S rDNA sequence for structural analysis of the nucleosome. These structures of an ancient nucleosome-binding motif suggest that there is an untapped realm of proteins with a related mode of binding to chromatin.
- Department of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, California 92093, USA.
Organizational Affiliation: