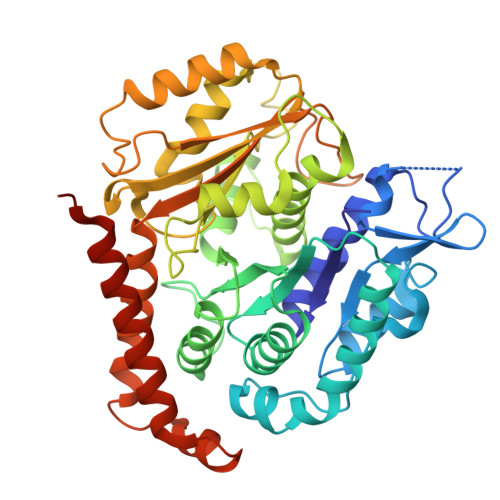
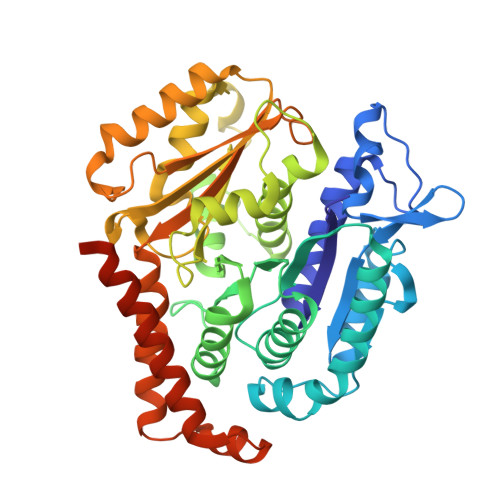
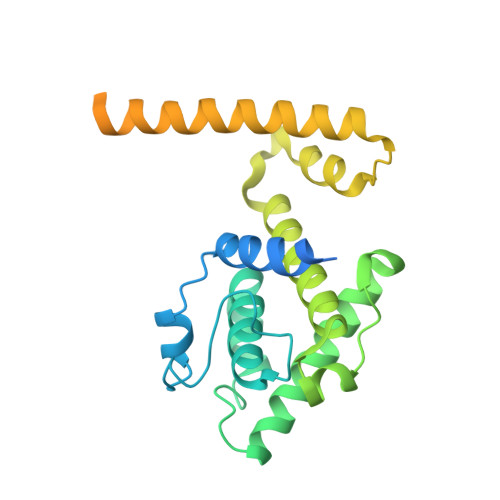
How augmin establishes the angle of the microtubule branch site.
Travis, S.M., Kraus, J., McManus, C.T., Golden, K., Zhang, R., Petry, S.(2025) Nat Commun 16: 9646-9646
- PubMed: 41173848
- DOI: https://doi.org/10.1038/s41467-025-64650-1
- Primary Citation of Related Structures:
9D2B, 9OLH - PubMed Abstract:
How microtubules (MTs) are generated in the proper orientation is essential to understanding how the cytoskeleton organizes a cell and MT-dependent events such as cell division. In the spindle, most MTs are generated through the branching MT nucleation pathway. In this pathway, new MTs are nucleated from the side of existing MTs and oriented at a shallow angle by the branching factor augmin, ensuring that both MTs have the same polarity. Yet, how augmin binds MTs and sets the branch angle has remained unclear. Here, we report the cryo-electron microscopy structure of an augmin subcomplex on the MT. This structure resembles that of NDC80 bound to the MT, with the conserved CH domain of augmin's Haus6 subunit directly proximal to the MT lattice. We find that the Haus6 CH domain is a bona fide MT binding site that increases augmin's affinity for the MT and helps establish branch angle. A second binding site, located in the disordered N-terminus of Haus8, also establishes branch angle,. Thus, we find that augmin regulates MT branching using two domains, each tuned to modulate MT affinity and MT branch angle. This work expands our mechanistic understanding of branching MT nucleation and thus spindle formation.
- Department of Molecular Biology, Princeton University, Princeton, NJ, USA.
Organizational Affiliation: