Antibacterial macrocyclic peptides reveal a distinct mode of BamA inhibition.
Walker, M.E., Zhu, W., Peterson, J.H., Wang, H., Patteson, J., Soriano, A., Zhang, H., Mayhood, T., Hou, Y., Mesbahi-Vasey, S., Gu, M., Frost, J., Lu, J., Johnston, J., Hipolito, C., Lin, S., Painter, R.E., Klein, D., Walji, A., Weinglass, A., Kelly, T.M., Saldanha, A., Schubert, J., Bernstein, H.D., Walker, S.S.(2025) Nat Commun 16: 3395-3395
- PubMed: 40210867
- DOI: https://doi.org/10.1038/s41467-025-58086-w
- Primary Citation of Related Structures:
9CS0, 9CS1, 9CS2 - PubMed Abstract:
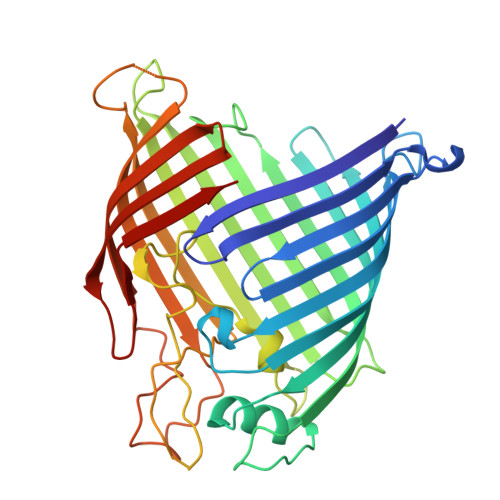
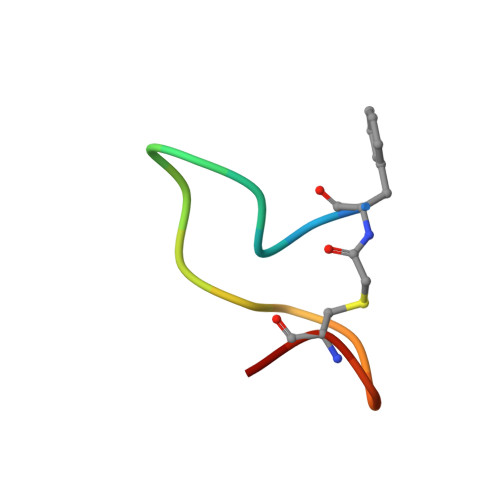
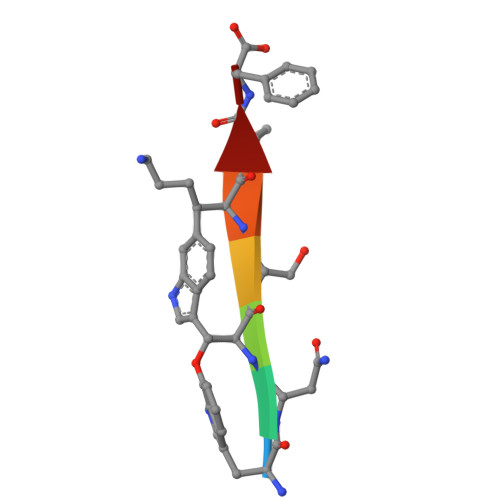
Outer membrane proteins (OMPs) produced by Gram-negative bacteria contain a cylindrical amphipathic β-sheet ("β-barrel") that functions as a membrane spanning domain. The assembly (folding and membrane insertion) of OMPs is mediated by the heterooligomeric β-barrel assembly machine (BAM). The central BAM subunit (BamA) is an attractive antibacterial target because its structure and cell surface localization are conserved, it catalyzes an essential reaction, and potent bactericidal compounds that inhibit its activity have been described. Here we utilize mRNA display to discover cyclic peptides that bind to Escherichia coli BamA with high affinity. We describe three peptides that arrest the growth of BAM deficient E. coli strains, inhibit OMP assembly in live cells and in vitro, and bind to unique sites within the BamA β-barrel lumen. Remarkably, we find that if the peptides are added to cultures after a slowly assembling OMP mutant binds to BamA, they accelerate its biogenesis. The data strongly suggest that the peptides trap BamA in conformations that block the initiation of OMP assembly but favor a later assembly step. Molecular dynamics simulations provide further evidence that the peptides bind stably to BamA and function by a previously undescribed mechanism.
- Merck & Co., Inc., West Point, PA, USA.
Organizational Affiliation: