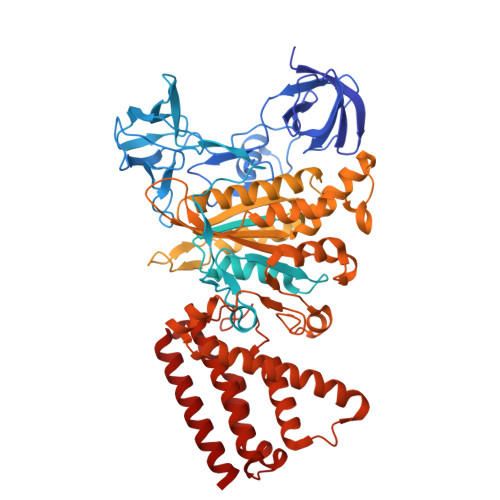
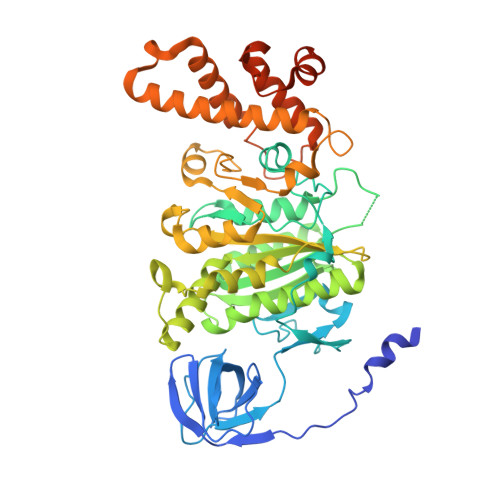
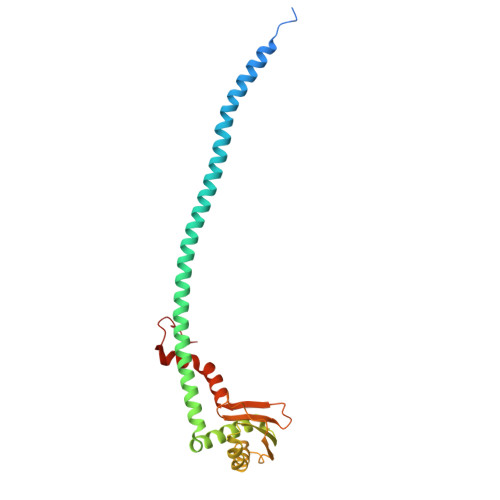
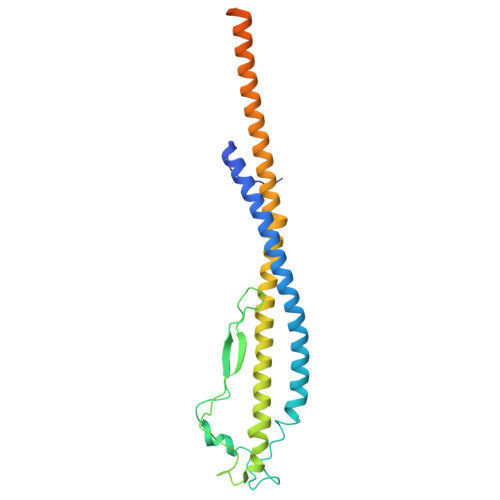
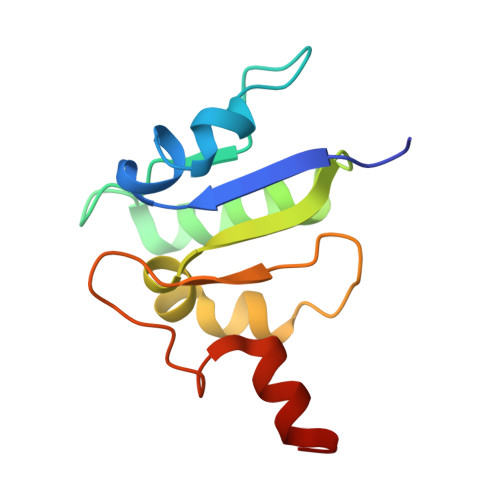
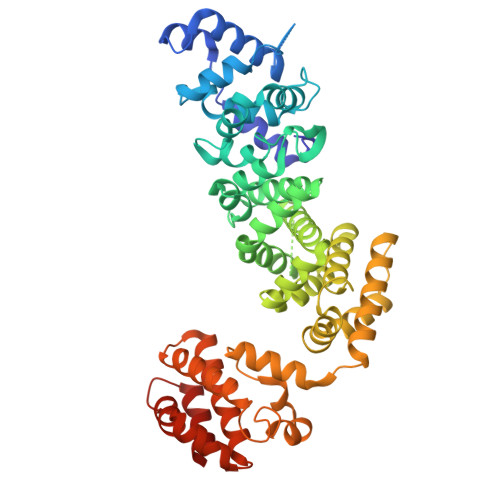
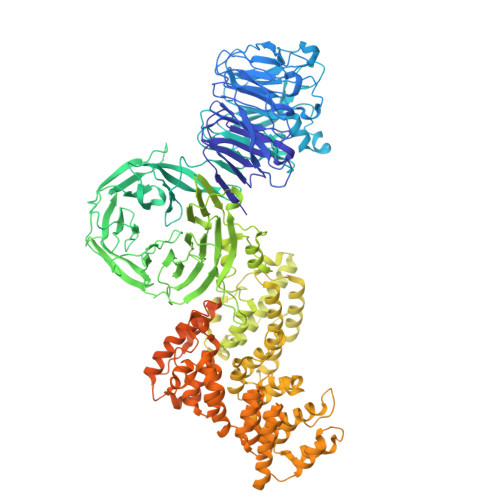
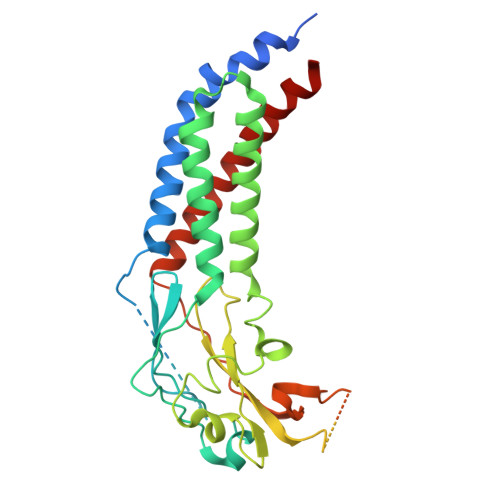
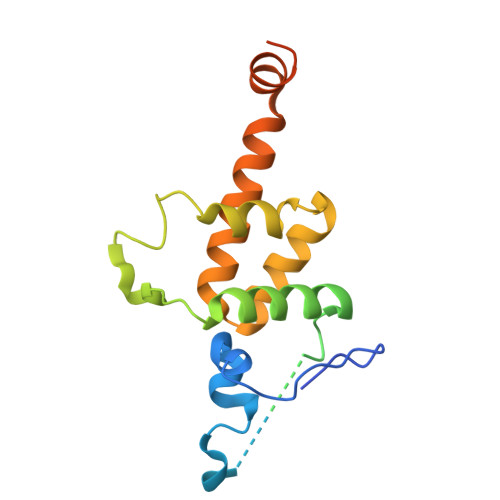
Structure of yeast RAVE bound to a partial V 1 complex.
Wang, H., Tarsio, M., Kane, P.M., Rubinstein, J.L.(2024) Proc Natl Acad Sci U S A 121: e2414511121-e2414511121
- PubMed: 39625975
- DOI: https://doi.org/10.1073/pnas.2414511121
- Primary Citation of Related Structures:
9COP - PubMed Abstract:
Vacuolar-type ATPases (V-ATPases) are membrane-embedded proton pumps that acidify intracellular compartments in almost all eukaryotic cells. Homologous with ATP synthases, these multisubunit enzymes consist of a soluble catalytic V 1 subcomplex and a membrane-embedded proton-translocating V O subcomplex. The V 1 and V O subcomplexes can undergo reversible dissociation to regulate proton pumping, with reassociation of V 1 and V O requiring the protein complex known as RAVE (regulator of the ATPase of vacuoles and endosomes). In the yeast Saccharomyces cerevisiae , RAVE consists of subunits Rav1p, Rav2p, and Skp1p. We used electron cryomicroscopy (cryo-EM) to determine a structure of yeast RAVE bound to V 1 . In the structure, RAVE is an L-shaped complex with Rav2p pointing toward the membrane and Skp1p distant from both the membrane and V 1 . Only Rav1p interacts with V 1 , binding to a region of subunit A not found in the corresponding ATP synthase subunit. When bound to RAVE, V 1 is in a rotational state suitable for binding the free V O complex, but in the structure, it is partially disrupted, missing five of its 16 subunits. Other than these missing subunits and the conformation of the inhibitory subunit H, the V 1 complex with RAVE appears poised for reassembly with V O .
- Molecular Medicine Program, The Hospital for Sick Children, Toronto, ON M5G 0A4, Canada.
Organizational Affiliation: