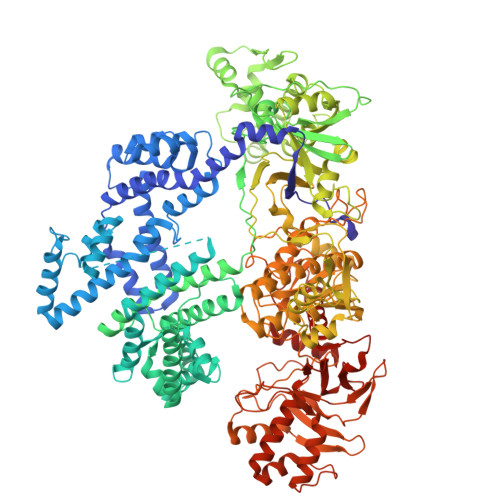
CRISPR-Cas12a bends DNA to destabilize base pairs during target interrogation.
Soczek, K.M., Cofsky, J.C., Tuck, O.T., Shi, H., Doudna, J.A.(2025) Nucleic Acids Res 53
- PubMed: 39698811
- DOI: https://doi.org/10.1093/nar/gkae1192
- Primary Citation of Related Structures:
9CJH, 9CJI, 9CJJ - PubMed Abstract:
RNA-guided endonucleases are involved in processes ranging from adaptive immunity to site-specific transposition and have revolutionized genome editing. CRISPR-Cas9, -Cas12 and related proteins use guide RNAs to recognize ∼20-nucleotide target sites within genomic DNA by mechanisms that are not yet fully understood. We used structural and biochemical methods to assess early steps in DNA recognition by Cas12a protein-guide RNA complexes. We show here that Cas12a initiates DNA target recognition by bending DNA to induce transient nucleotide flipping that exposes nucleobases for DNA-RNA hybridization. Cryo-EM structural analysis of a trapped Cas12a-RNA-DNA surveillance complex and fluorescence-based conformational probing show that Cas12a-induced DNA helix destabilization enables target discovery and engagement. This mechanism of initial DNA interrogation resembles that of CRISPR-Cas9 despite distinct evolutionary origins and different RNA-DNA hybridization directionality of these enzyme families. Our findings support a model in which RNA-mediated DNA interference begins with local helix distortion by transient CRISPR-Cas protein binding.
- Department of Molecular and Cell Biology, University of California Berkeley, Berkeley, CA, USA.
Organizational Affiliation: