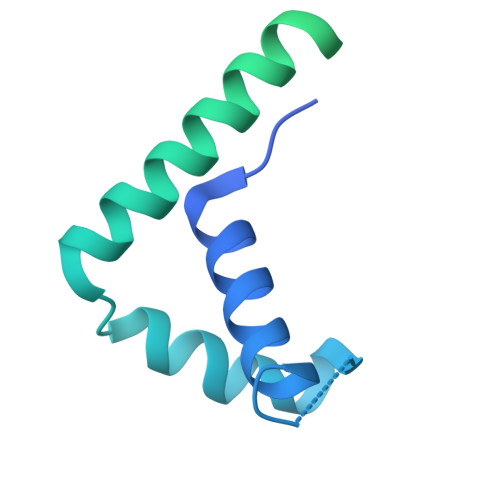
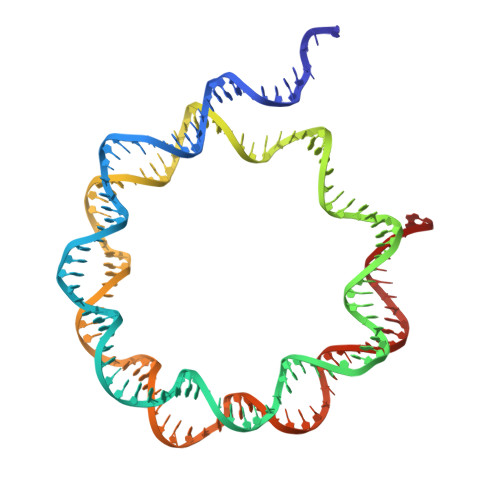
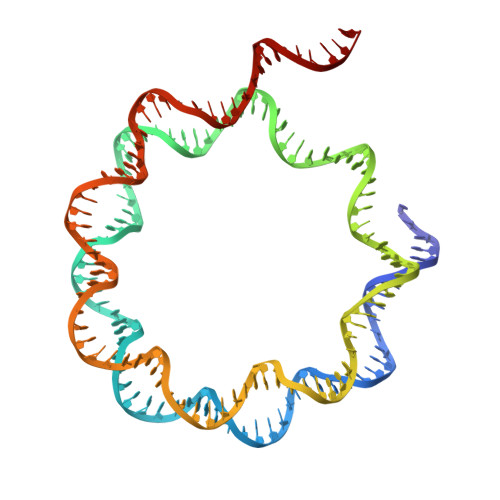
HMGB1 deforms nucleosomal DNA to generate a dynamic chromatin environment counteracting the effects of linker histone.
Saunders, H.S., Chio, U.S., Moore, C.M., Ramani, V., Cheng, Y., Narlikar, G.J.(2025) Sci Adv 11: eads4473-eads4473
- PubMed: 40815652
- DOI: https://doi.org/10.1126/sciadv.ads4473
- Primary Citation of Related Structures:
9CG9 - PubMed Abstract:
The essential architectural protein HMGB1 increases accessibility of nucleosomal DNA and counteracts the effects of linker histone H1. However, HMGB1 is less abundant than H1 and binds nucleosomes more weakly, raising the question of how it competes with H1. Here, we find that HMGB1 increases nucleosomal DNA accessibility without displacing H1. HMGB1 also increases the dynamics of condensed, H1-bound chromatin. Unexpectedly, cryo-electron microscopy structures show HMGB1 bound at internal locations on nucleosomes and local DNA distortion. These sites are away from where H1 binds, explaining how HMGB1 and H1 can co-occupy a nucleosome. Our findings suggest a model where HMGB1 counteracts the effects of H1 by distorting nucleosomal DNA and disrupting interactions of the H1 carboxyl-terminal tail with DNA. Compared to mutually exclusive binding, co-occupancy by HMGB1 and H1 allows greater diversity in dynamic chromatin states. More generally, these results explain how architectural proteins acting at the nucleosome scale can have large effects on chromatin dynamics at the mesoscale.
- Department of Biochemistry and Biophysics, University of California, San Francisco, San Francisco, CA 94158, USA.
Organizational Affiliation: