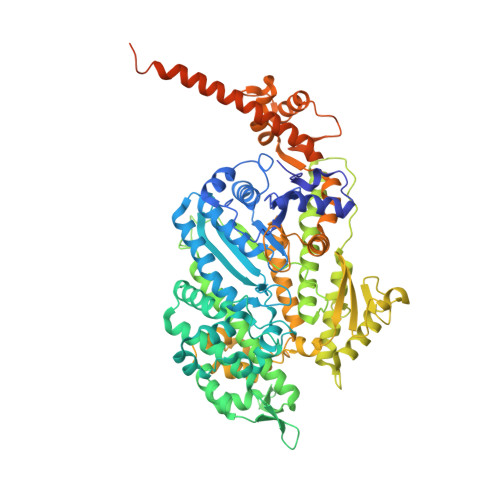
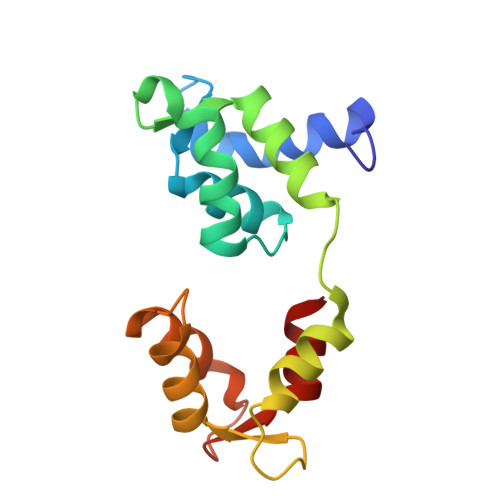
High-resolution structures of Myosin-IC reveal a unique actin-binding orientation, ADP release pathway, and power stroke trajectory.
Chavali, S.S., Carman, P.J., Shuman, H., Ostap, E.M., Sindelar, C.V.(2025) Proc Natl Acad Sci U S A 122: e2415457122-e2415457122
- PubMed: 40014570
- DOI: https://doi.org/10.1073/pnas.2415457122
- Primary Citation of Related Structures:
9CFU, 9CFV, 9CFW, 9CFX - PubMed Abstract:
Myosin-IC (myo1c) is a class-I myosin that supports transport and remodeling of the plasma membrane and membrane-bound vesicles. Like other members of the myosin family, its biochemical kinetics are altered in response to changes in mechanical loads that resist the power stroke. However, myo1c is unique in that the primary force-sensitive kinetic transition is the isomerization that follows ATP binding, not ADP release as in other slow myosins. Myo1c also powers actin gliding along curved paths, propelling actin filaments in leftward circles. To understand the origins of this unique force-sensing and motile behavior, we solved actin-bound myo1c cryo-EM structures in the presence and absence of ADP. Our structures reveal that in contrast with other myosins, the myo1c lever arm swing is skewed, partly due to a different actin interface that reorients the motor domain on actin. The structures also reveal unique nucleotide-dependent behavior of both the nucleotide pocket as well as an element called the N-terminal extension (NTE). We incorporate these observations into a model that explains why force primarily regulates ATP binding in myo1c, rather than ADP release as in other myosins. Integrating our cryo-EM data with available crystallography structures allows the modeling of full-length myo1c during force generation, supplying insights into its role in membrane remodeling. These results highlight how relatively minor sequence differences in members of the myosin superfamily can significantly alter power stroke geometry and force-sensing properties, with important implications for biological function.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520-8103.
Organizational Affiliation: