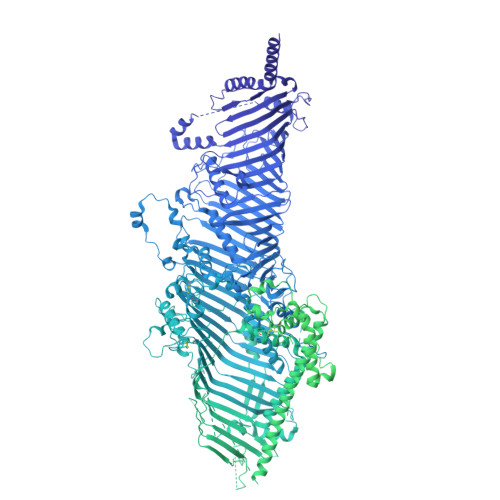
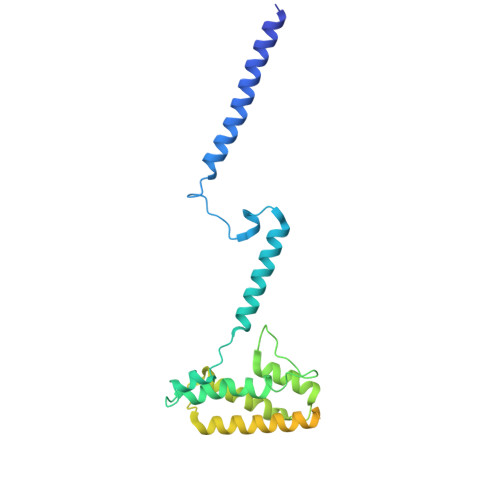
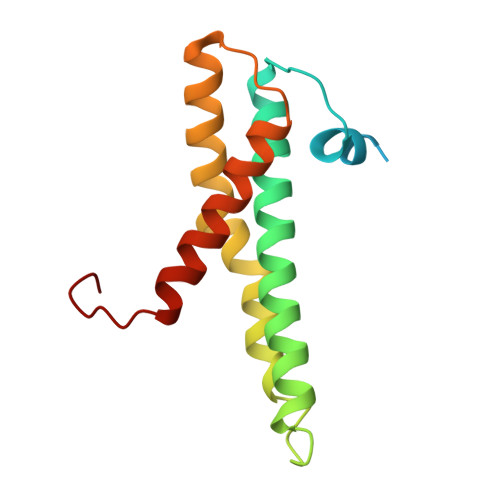
Structural basis of lipid transfer by a bridge-like lipid-transfer protein.
Kang, Y., Lehmann, K.S., Long, H., Jefferson, A., Purice, M., Freeman, M., Clark, S.(2025) Nature 642: 242-249
- PubMed: 40269155
- DOI: https://doi.org/10.1038/s41586-025-08918-y
- Primary Citation of Related Structures:
9CAP - PubMed Abstract:
Bridge-like lipid-transport proteins (BLTPs) are an evolutionarily conserved family of proteins that localize to membrane-contact sites and are thought to mediate the bulk transfer of lipids from a donor membrane, typically the endoplasmic reticulum, to an acceptor membrane, such as that of the cell or an organelle 1 . Although BLTPs are fundamentally important for a wide array of cellular functions, their architecture, composition and lipid-transfer mechanisms remain poorly characterized. Here we present the subunit composition and the cryogenic electron microscopy structure of the native LPD-3 BLTP complex isolated from transgenic Caenorhabditis elegans. LPD-3 folds into an elongated, rod-shaped tunnel of which the interior is filled with ordered lipid molecules that are coordinated by a track of ionizable residues that line one side of the tunnel. LPD-3 forms a complex with two previously uncharacterized proteins, one of which we have named Spigot and the other of which remains unnamed. Spigot interacts with the N-terminal end of LPD-3 where lipids are expected to enter the tunnel, and experiments in multiple model systems indicate that Spigot has a conserved role in BLTP function. Our LPD-3 complex structural data reveal protein-lipid interactions that suggest a model for how the native LPD-3 complex mediates bulk lipid transport and provides a foundation for mechanistic studies of BLTPs.
- Vollum Institute, Oregon Health & Science University, Portland, OR, USA.
Organizational Affiliation: