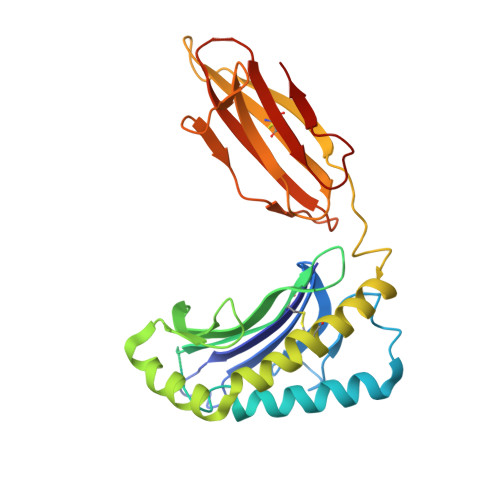
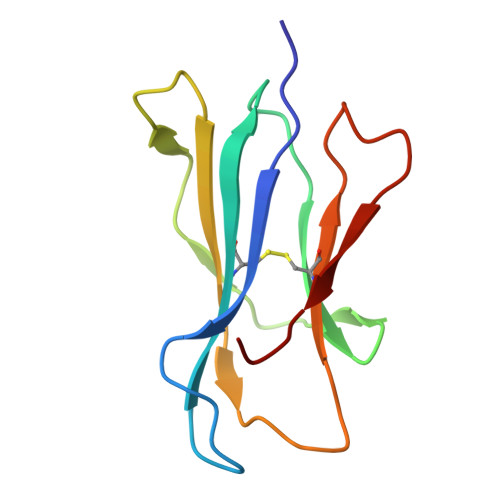
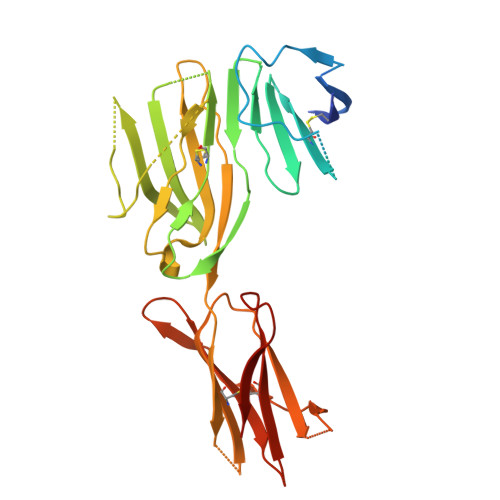
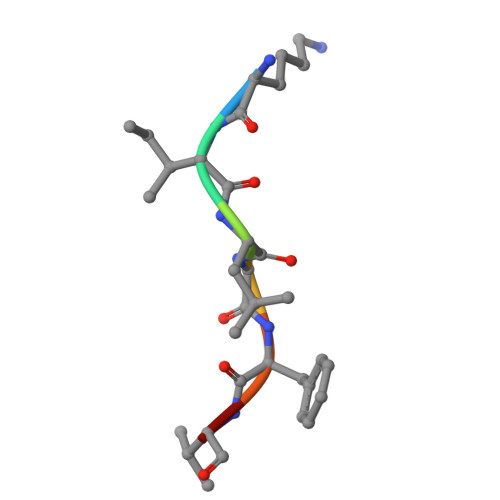
CryoEM structure of an MHC-I/TAPBPR peptide-bound intermediate reveals the mechanism of antigen proofreading.
Sun, Y., Pumroy, R.A., Mallik, L., Chaudhuri, A., Wang, C., Hwang, D., Danon, J.N., Dasteh Goli, K., Moiseenkova-Bell, V.Y., Sgourakis, N.G.(2025) Proc Natl Acad Sci U S A 122: e2416992122-e2416992122
- PubMed: 39786927
- DOI: https://doi.org/10.1073/pnas.2416992122
- Primary Citation of Related Structures:
9C96 - PubMed Abstract:
Class I major histocompatibility complex (MHC-I) proteins play a pivotal role in adaptive immunity by displaying epitopic peptides to CD8+ T cells. The chaperones tapasin and TAPBPR promote the selection of immunogenic antigens from a large pool of intracellular peptides. Interactions of chaperoned MHC-I molecules with incoming peptides are transient in nature, and as a result, the precise antigen proofreading mechanism remains elusive. Here, we leverage a high-fidelity TAPBPR variant and conformationally stabilized MHC-I, to determine the solution structure of the human antigen editing complex bound to a peptide decoy by cryogenic electron microscopy (cryo-EM) at an average resolution of 3.0 Å. Antigen proofreading is mediated by transient interactions formed between the nascent peptide binding groove with the P2/P3 peptide anchors, where conserved MHC-I residues stabilize incoming peptides through backbone-focused contacts. Finally, using our high-fidelity chaperone, we demonstrate robust peptide exchange on the cell surface across multiple clinically relevant human MHC-I allomorphs. Our work has important ramifications for understanding the selection of immunogenic epitopes for T cell screening and vaccine design applications.
- Department of Biochemistry and Biophysics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA 19104.
Organizational Affiliation: