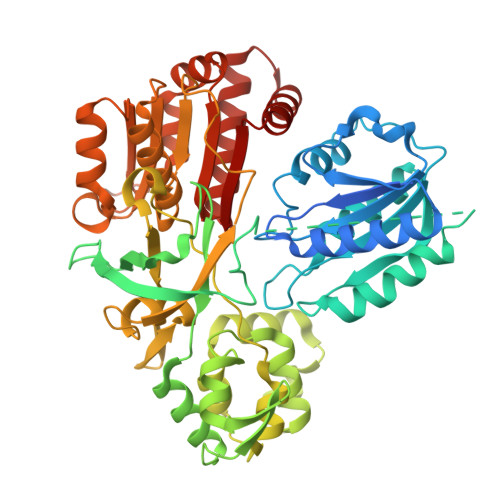
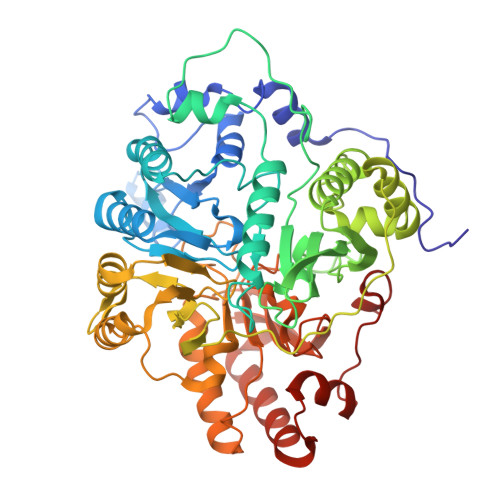
Structure of dimerized assimilatory NADPH-dependent sulfite reductase reveals the minimal interface for diflavin reductase binding.
Ghazi Esfahani, B., Walia, N., Neselu, K., Garg, Y., Aragon, M., Askenasy, I., Wei, H.A., Mendez, J.H., Stroupe, M.E.(2025) Nat Commun 16: 2955-2955
- PubMed: 40140349
- DOI: https://doi.org/10.1038/s41467-025-58037-5
- Primary Citation of Related Structures:
9C91 - PubMed Abstract:
Escherichia coli NADPH-dependent assimilatory sulfite reductase (SiR) reduces sulfite by six electrons to make sulfide for incorporation into sulfur-containing biomolecules. SiR has two subunits: an NADPH, FMN, and FAD-binding diflavin flavoprotein and a siroheme/Fe 4 S 4 cluster-containing hemoprotein. The molecular interactions that govern subunit binding have been unknown since the discovery of SiR over 50 years ago because SiR is flexible, thus has been intransigent for traditional high-resolution structural analysis. We use a combination of the chameleon® plunging system with a fluorinated lipid to overcome the challenges of preserving a flexible molecule to determine a 2.78 Å-resolution cryo-EM structure of a minimal heterodimer complex. Chameleon®, combined with the fluorinated lipid, overcomes persistent denaturation at the air-water interface. Using a previously characterized minimal heterodimer reduces the heterogeneity of a structurally heterogeneous complex to a level that we analyze using multi-conformer cryo-EM image analysis algorithms. Here, we report the near-atomic resolution structure of the flavoprotein/hemoprotein complex, revealing how they interact in a minimal interface. Further, we determine the structural elements that discriminate between pairing a hemoprotein with a diflavin reductase, as in the E. coli homolog, or a ferredoxin partner, as in maize (Zea mays).
- Department of Biological Science and Institute of Molecular Biophysics, Florida State University, Tallahassee, FL, USA.
Organizational Affiliation: