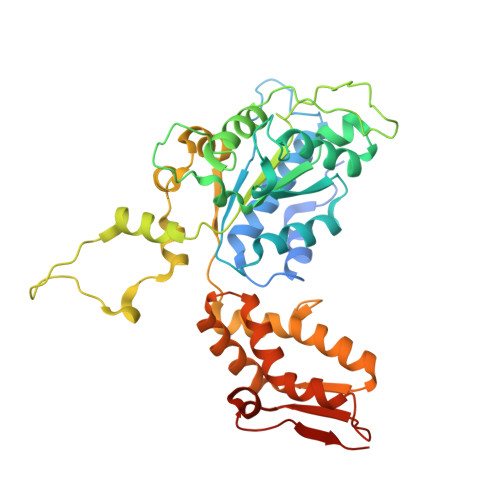
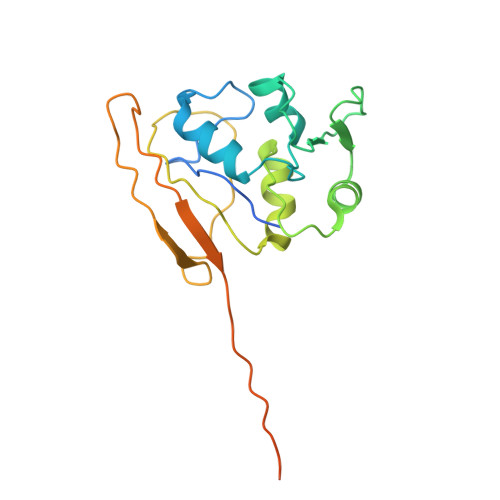
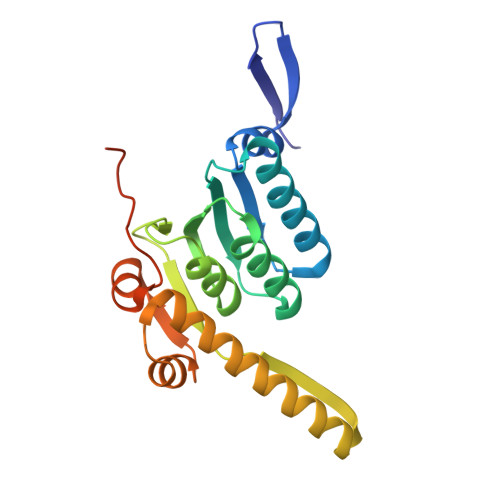
A proteolytic AAA+ machine poised to unfold protein substrates.
Ghanbarpour, A., Sauer, R.T., Davis, J.H.(2024) Nat Commun 15: 9681-9681
- PubMed: 39516482
- DOI: https://doi.org/10.1038/s41467-024-53681-9
- Primary Citation of Related Structures:
8V9R, 9C87, 9C88 - PubMed Abstract:
AAA+ proteolytic machines unfold proteins before degrading them. Here, we present cryoEM structures of ClpXP-substrate complexes that reveal a postulated but heretofore unseen intermediate in substrate unfolding/degradation. A ClpX hexamer draws natively folded substrates tightly against its axial channel via interactions with a fused C-terminal degron tail and ClpX-RKH loops that flexibly conform to the globular substrate. The specific ClpX-substrate contacts observed vary depending on the substrate degron and affinity tags, helping to explain ClpXP's ability to unfold/degrade a wide array of different cellular substrates. Some ClpX contacts with native substrates are enabled by upward movement of the seam subunit in the AAA+ spiral, a motion coupled to a rearrangement of contacts between the ClpX unfoldase and ClpP peptidase. Our structures additionally highlight ClpX's ability to translocate a diverse array of substrate topologies, including the co-translocation of two polypeptide chains.
- Department of Biochemistry and Molecular Biophysics, Washington University in St. Louis, St Louis, 63130, USA.
Organizational Affiliation: