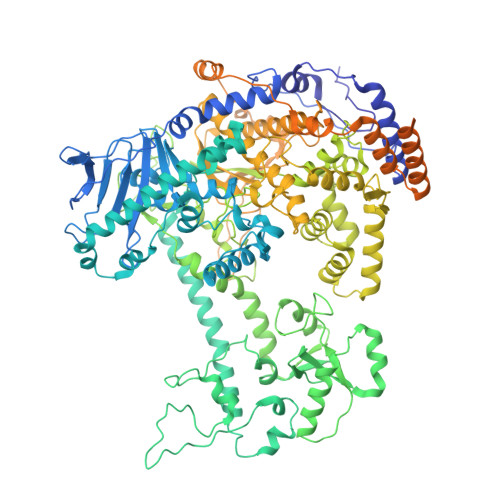
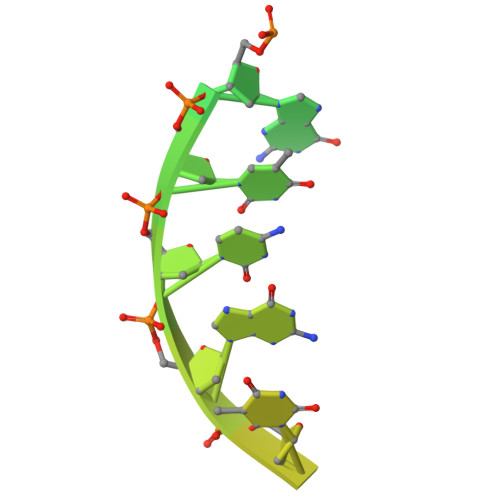
Structural basis for intrinsic strand displacement activity of mitochondrial DNA polymerase.
Nayak, A.R., Sokolova, V., Sillamaa, S., Herbine, K., Sedman, J., Temiakov, D.(2025) Nat Commun 16: 2417-2417
- PubMed: 40069189
- DOI: https://doi.org/10.1038/s41467-025-57594-z
- Primary Citation of Related Structures:
9C51, 9C52, 9C53, 9D2I - PubMed Abstract:
Members of the Pol A family of DNA polymerases, found across all domains of life, utilize various strategies for DNA strand separation during replication. In higher eukaryotes, mitochondrial DNA polymerase γ relies on the replicative helicase TWINKLE, whereas the yeast ortholog, Mip1, can unwind DNA independently. Using Mip1 as a model, we present a series of high-resolution cryo-EM structures that capture the process of DNA strand displacement. Our data reveal previously unidentified structural elements that facilitate the unwinding of the downstream DNA duplex. Yeast cells harboring Mip1 variants defective in strand displacement exhibit impaired oxidative phosphorylation and loss of mtDNA, corroborating the structural observations. This study provides a molecular basis for the intrinsic strand displacement activity of Mip1 and illuminates the distinct unwinding mechanisms utilized by Pol A family DNA polymerases.
- Department of Biochemistry and Molecular Biology, Thomas Jefferson University; 1020 Locust St, Philadelphia, USA.
Organizational Affiliation: