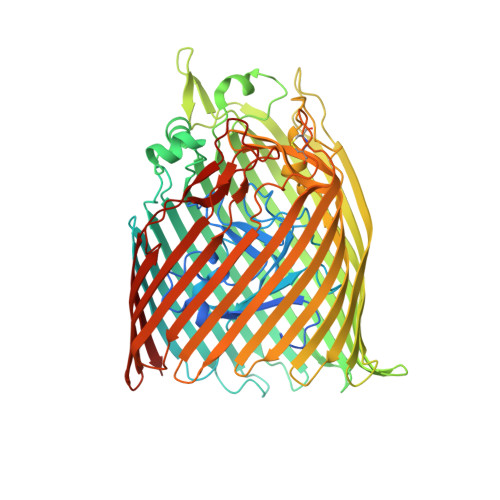
High-affinity PQQ import is widespread in Gram-negative bacteria.
Munder, F., Voutsinos, M., Hantke, K., Venugopal, H., Grinter, R.(2025) Sci Adv 11: eadr2753-eadr2753
- PubMed: 40446051
- DOI: https://doi.org/10.1126/sciadv.adr2753
- Primary Citation of Related Structures:
9C4O - PubMed Abstract:
Pyrroloquinoline quinone (PQQ) is a soluble redox cofactor used by diverse bacteria. Many Gram-negative bacteria that encode PQQ-dependent enzymes do not produce it and instead obtain it from the environment. To achieve this, Escherichia coli uses the TonB-dependent transporter PqqU as a high-affinity PQQ importer. Here, we show that PqqU binds PQQ with high affinity and determine the high-resolution structure of the PqqU-PQQ complex, revealing that PqqU undergoes conformational changes in PQQ binding to capture the cofactor in an internal cavity. We show that these conformational changes preclude the binding of a bacteriophage, which targets PqqU as a cell surface receptor. Guided by the PqqU-PQQ structure, we identify amino acids essential for PQQ import and leverage this information to map the presence of PqqU across Gram-negative bacteria. This reveals that PqqU is encoded by Gram-negative bacteria from at least 22 phyla occupying diverse habitats, indicating that PQQ is an important cofactor for bacteria that adopt diverse lifestyles and metabolic strategies.
- Department of Microbiology, Monash Biomedicine Discovery Institute, Monash University, Clayton, Australia.
Organizational Affiliation: