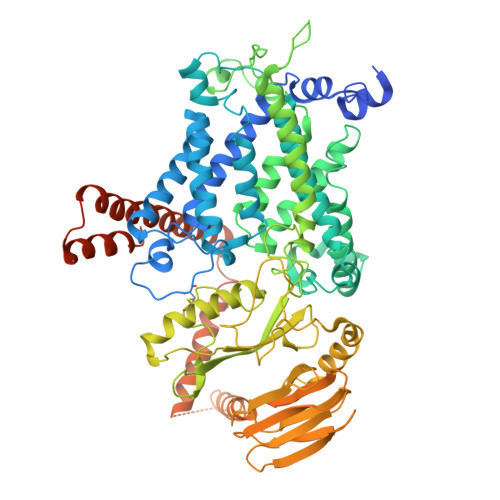
Structure and mechanism of vitamin-K-dependent gamma-glutamyl carboxylase.
Wang, R., Chen, B., Elghobashi-Meinhardt, N., Tie, J.K., Ayala, A., Zhou, N., Qi, X.(2025) Nature 639: 808-815
- PubMed: 39880952
- DOI: https://doi.org/10.1038/s41586-024-08484-9
- Primary Citation of Related Structures:
9BUM, 9BUR, 9BUX - PubMed Abstract:
γ-Glutamyl carboxylase (GGCX) is the sole identified enzyme that uses vitamin K (VK) as a cofactor in humans. This protein catalyses the oxidation of VK hydroquinone to convert specific glutamate residues to γ-carboxyglutamate residues in VK-dependent proteins (VDPs), which are involved in various essential biological processes and diseases 1-3 . However, the working mechanism of GGCX remains unclear. Here we report three cryogenic electron microscopy structures of human GGCX: in the apo state, bound to osteocalcin (a VDP) and bound to VK. The propeptide of the VDP binds to the lumenal domain of GGCX, which stabilizes transmembrane helices 6 and 7 of GGCX to create the VK-binding pocket. After binding of VK, residue Lys218 in GGCX mediates the oxidation of VK hydroxyquinone, which leads to the deprotonation of glutamate residues and the construction of γ-carboxyglutamate residues. Our structural observations and results from binding and cell biological assays and molecular dynamics simulations show that a cholesterol molecule interacts with the transmembrane helices of GGCX to regulate its protein levels in cells. Together, these results establish a link between cholesterol metabolism and VK-dependent pathways.
- Department of Molecular Genetics, University of Texas Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: