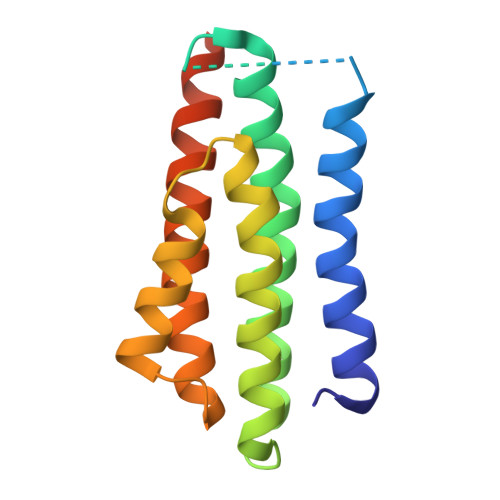
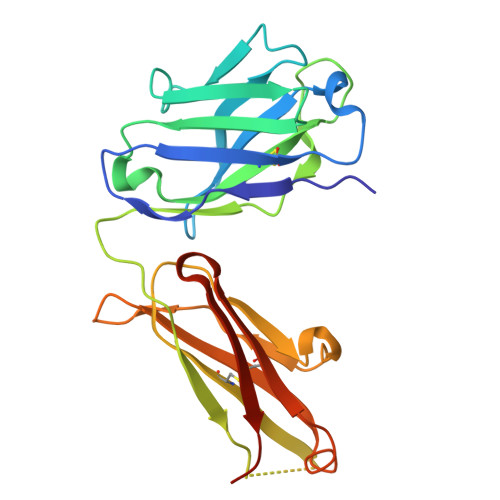
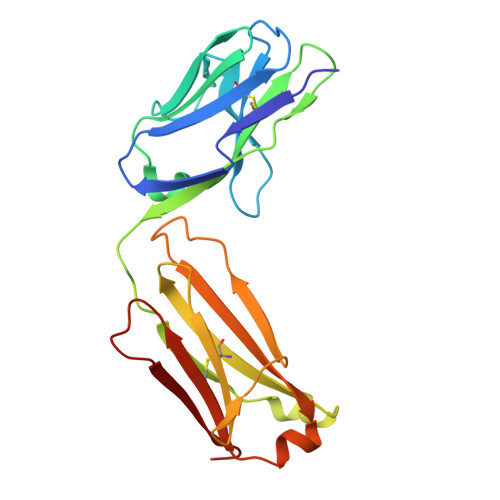
Germline encoded residues dominate the interaction of a human monoclonal antibody with decorin binding protein A of Borrelia burgdorferi.
Rudolph, M.J., Muriuki, B.M., Chen, Y., Vance, D.J., Vorauer, C., Piazza, C.L., Freeman-Gallant, G., Golonka, R.M., Mirabile, G., Guttman, M., Cavacini, L.A., Mantis, N.J.(2025) Front Immunol 16: 1611828-1611828
- PubMed: 40692775
- DOI: https://doi.org/10.3389/fimmu.2025.1611828
- Primary Citation of Related Structures:
9BQW - PubMed Abstract:
During the course of Lyme disease, humans mount a robust and sustained antibody response against dozens of Borrelia burgdorferi outer surface lipoproteins. Identifying which antibodies are associated with spirochete clearance and disease resolution is of paramount importance in therapeutic development. In this study, we describe the isolation and structural characterization of a human monoclonal antibody (MAb) against decorin binding protein A (DbpA), one of the most immunogenic of B. burgdorferi 's outer surface proteins. High-resolution epitope mapping by HDX-MS and X-ray crystallography revealed that F945 associates with a lateral face of DbpA in a side-on orientation without obstructing resides associated with DbpA's ability to bind components of the extracellular matrix. The structure of the DbpA-F945 Fab complex revealed an outsized role for variable light chain (V L ) germline encoded residues in mediating DbpA interactions. In fact, the majority of the critical contacts between F945 and DbpA involved V k 1-33 germline encoded residues, suggesting that certain human B cell receptors (BCR) may be preconfigured to recognize DbpA and therefore have a lower threshold for B cell activation and clonal development. Passive administration of F945 IgG was not sufficient to protect against B. burgdorferi in a mouse model of needle infection, although these experiments do not rule out a role for F945 in influencing B. burgdorferi tissue tropism and/or retention within specific niches. Nonetheless, it is tempting to speculate that F945 represents a class of DbpA antibodies with value in Lyme disease diagnostics, but that may not contribute to B. burgdorferi clearance or disease resolution in humans.
- New York Structural Biology Center, New York, NY, United States.
Organizational Affiliation: