Structural studies of the IFN lambda 4 receptor complex using cryoEM enabled by protein engineering.
Grubbe, W.S., Zhang, B., Kauffman, A., Bylehn, F., Padol, K., Jung, H.G., Park, S.B., Priest, J.M., Ozkan, E., de Pablo, J.J., Liang, T.J., Zhao, M., Mendoza, J.L.(2025) Nat Commun 16: 818-818
- PubMed: 39827213
- DOI: https://doi.org/10.1038/s41467-025-56119-y
- Primary Citation of Related Structures:
9BPU, 9BPV - PubMed Abstract:
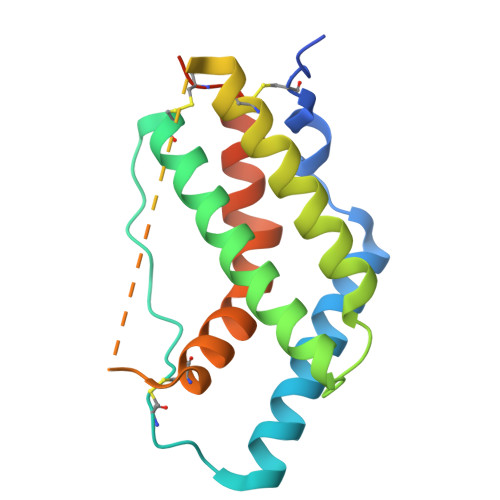
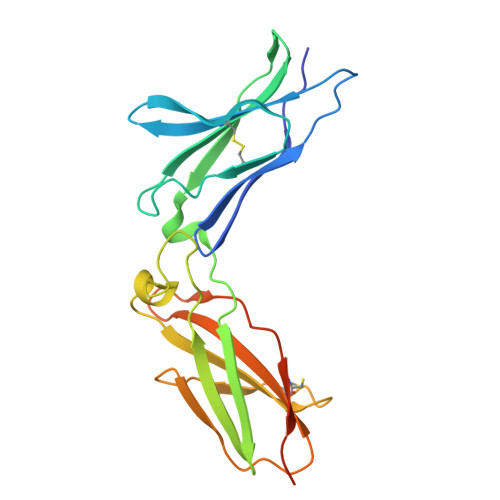
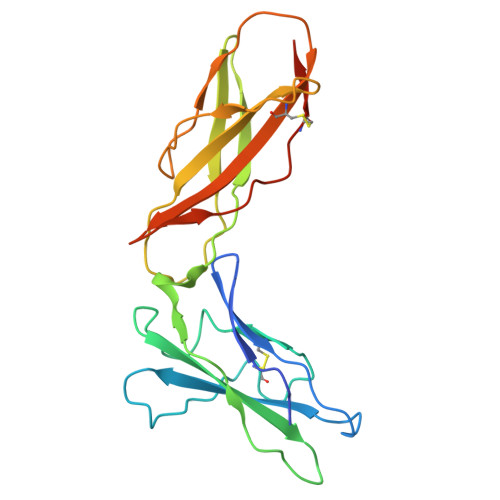
IFNλ4 has posed a conundrum in human immunology since its discovery in 2013, with its expression linked to complications with viral clearance. While genetic and cellular studies revealed the detrimental effects of IFNλ4 expression, extensive structural and functional characterization has been limited by the inability to express and purify the protein, complicating explanations of its paradoxical behavior. In this work, we report a method for robust production of IFNλ4. We then use yeast surface display to affinity-mature IL10Rβ and solve the 72 kilodalton structures of IFNλ4 (3.26 Å) and IFNλ3 (3.00 Å) in complex with their receptors IFNλR1 and IL10Rβ using cryogenic electron microscopy. Comparison of the structures highlights differences in receptor engagement and reveals a distinct 12-degree rotation in overall receptor geometry, providing a potential mechanistic explanation for differences in cell signaling, downstream gene induction, and antiviral activities. Further, we perform a structural analysis using molecular modeling and simulation to identify a unique region of IFNλ4 that, when replaced, enables secretion of the protein from cells. These findings provide a structural and functional understanding of the IFNλ4 protein and enable future comprehensive studies towards correcting IFNλ4 dysfunction in large populations of affected patients.
- Pritzker School of Molecular Engineering, University of Chicago, Chicago, IL, USA.
Organizational Affiliation: